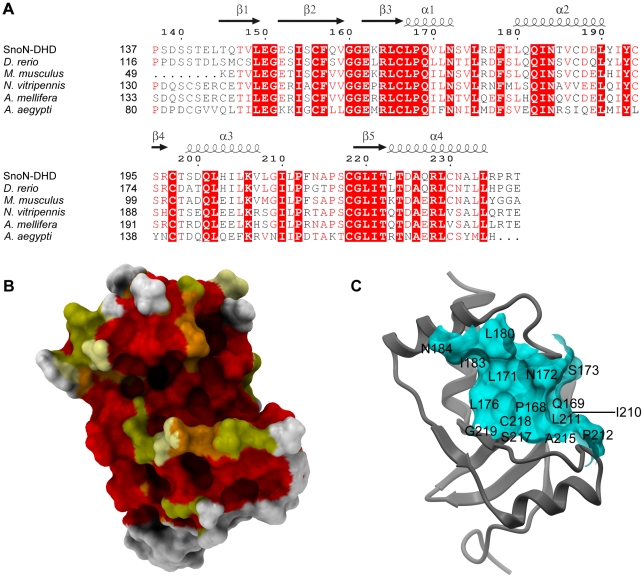
Figure 3. Conserved regions in SnoN-DHD.
Panel A: Sequence alignment between human SnoN-DHD and five orthologous proteins. The sequence of SnoN-DHD corresponds to the protein fragment used in structural determination. The Genbank ID of the five orthologous sequences are: Danio rerio (gi:190338009), Mus musculus (gi:113205055), Nasonia vitripennis (gi: 156550231), Apis mellifera (gi: 110748778), and Aedes aegypti (gi: 157124676). Secondary structure elements and numbering of SnoN-DHD are depicted above the alignment. Alignment was generated by ClustalW [41] and colored by ESPript [40]. Panel B: Conserved residues on the surface of SnoN-DHD. The molecular surface of SnoN-DHD is colored according to the sequence conservation from the alignment in Fig 3A. Residues are colored in red and orange if they are strictly conserved in six or five sequences, respectively. Yellow and pale yellow represent residues that are similar in six and five sequences, respectively. Other residues are colored in white. SnoN-DHD is in the same orientation as in Fig 2A. Panel C: Conserved groove on the surface of SnoN-DHD. Structure of SnoN-DHD is represented in ribbon mode. The molecular surface of the conserved groove mentioned in the text is displayed and colored in light blue. It is composed of the following residues: Leu167, Pro168, Gln169, Leu171, Asn172, Ser173, Leu 180, Ile183, Asn184, Ile210, Leu211, Pro212, Ala215, Ser217, Cys218 and Gly219. The position of these residues on the surface is indicated by their corresponding one-letter label.