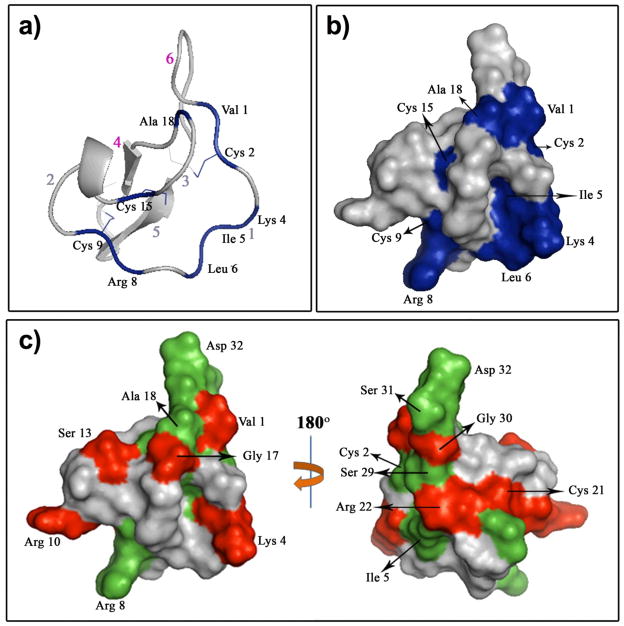
Figure 2.
Trypsin binding to MCoTI-I affects the MCoTI-I backbone dynamics. Ribbon (a) and surface (b) diagrams of the trypsin-MCoTI-I interaction map. Red arabic numbers indicate the positions of the MCoTI-I loops. The MCoTI-I residues with a large chemical shift difference (>0.3 ppm) are in blue. (c) Changes in the MCoTI-I order parameter due to binding to trypsin. Residues with Sf2 - Sb2 > 0.2, where Sf/b2 is the order parameter of the free or trypsin bound MCoTI-I, respectively, are depicted in red. MCoTI-I residues that were broadened in {15N,1H}-HSQC due to binding to trypsin are shown in green. We used a structure of free MCoTI-II (PDB code: 1IB9)[6] to illustrate the changes of MCoTI-I dynamics due to trypsin binding.