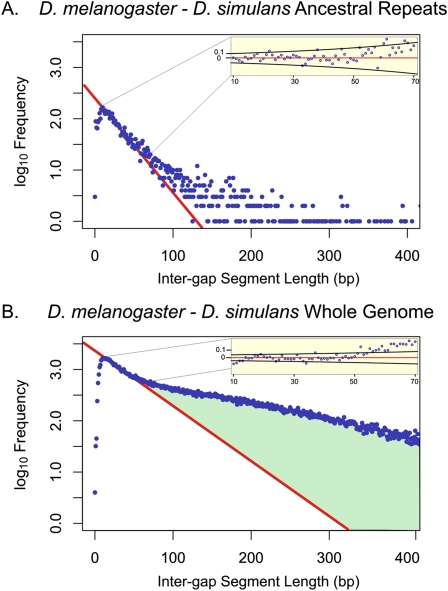
Figure 4.
Representative genomic distribution of IGS lengths in D. melanogaster and D. simulans alignments Frequencies of IGS (blue) lengths shown on a log10 scale for AR regions (A) and whole-genome sequences (B) with a G+C content of 0.495–0.445. The predictions of the neutral indel model are shown in red. In contrast to AR sequence for mouse and rat (Fig. 1), a relatively large proportion (20%–23%) of the small number of ancient fruit fly transposons appear to be under constraint, although the absolute quantity of sequence remains low (0.29–0.32 Mb). Similarly, for whole-genome sequence, we estimate that 55.5–66.2 Mb (46%–55%) of the genome is subject to constraint regarding indels. The difference in the predictions of the neutral indel model for whole-genome and AR sequence indicates that functional sequence may contribute to Drosophila short IGS.