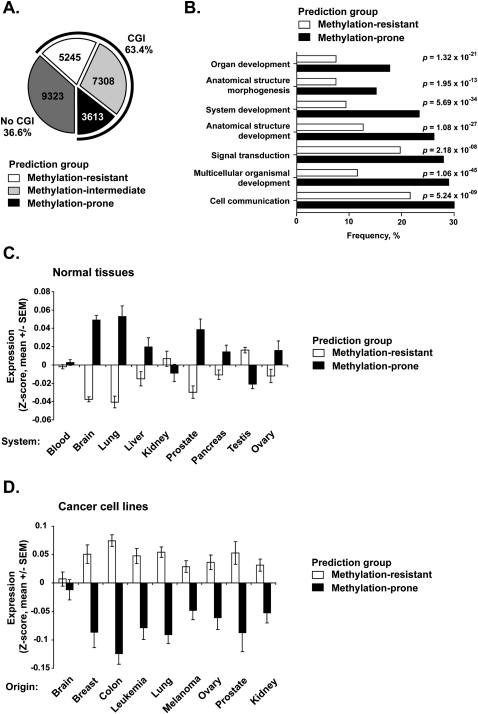
Figure 5.
Genome-wide prediction of predisposition to DNA methylation in cancer. (A) The pie chart shows the number of RefSeq genes with no CpG islands (dark gray) and the number of predicted methylation-resistant (white), methylation-intermediate (light gray), and methylation-prone (black) genes in promoter CpG island genes. (B) Gene Ontology (GO) analysis of 1952 predicted methylation-prone and 2583 predicted methylation-resistant genes for which functional information was available. Horizontal bars represent the frequency of significant GO terms. (C) Gene expression analysis for 2822 promoter CpG island associated genes predicted methylation-prone and 3651 predicted methylation-resistant genes in normal tissues. Expression values were retrieved from the GNF database (Su et al. 2004) and Z-score normalized per tissue. Each bar represents the mean ± SEM expression values in each tissue according to their predicted methylation predisposition. (D) Gene expression analysis for 599 promoter CpG island associated genes predicted methylation-prone and 996 predicted methylation-resistant genes in 52 cancer cell lines. Expression values were retrieved from a published work (Ross et al. 2000) and were analyzed as described in C. Only genes present in the studied array platforms could be evaluated, resulting in a different number of analyzed genes in each experiment.