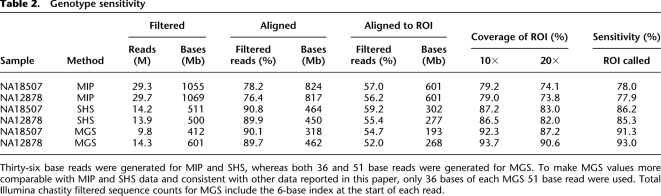
Table 2.
Genotype sensitivity
Thirty-six base reads were generated for MIP and SHS, whereas both 36 and 51 base reads were generated for MGS. To make MGS values more comparable with MIP and SHS data and consistent with other data reported in this paper, only 36 bases of each MGS 51 base read were used. Total Illumina chastity filtered sequence counts for MGS include the 6-base index at the start of each read.