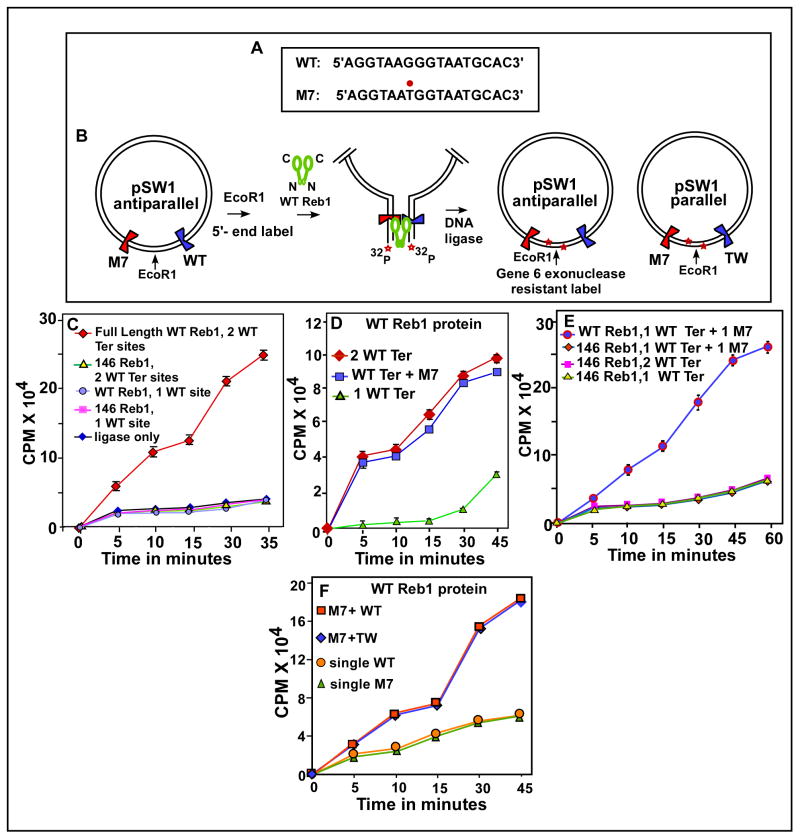
Fig. 2. In vitro Reb1-mediated ligation enhancement assay showing that Reb1 promotes interactions between not only two WT Ter sites but also between a WT and a nonfunctional mutant Ter site M7 in vitro.
A, sequences showing the WT, canonical Ter and the M7 mutant form with the G to T transversion marked with a red dot; B, a schematic representation of the assay method, the plasmid pSW1 contains either two WT Ter sites or a WT and a M7 site placed on either sides of the unique EcoR1 site, DNA looping caused by the binding of the WT Reb1 but not a truncated, monomeric mutant for is manifested by enhancement of DNA circularization of the linearized, labeled DNA that is measured by counting gene 6 exonuclease resistant label; C, Ligation enhancement kinetics showing that two WT sites loop DNA in the presence of the WT Reb1 but not when the truncated form of the protein was supplied, controls show no enhancement of ligation kinetics when a single Ter site was incubated with either the WT or the truncated form of Reb1 or when only ligase was added to the reaction mixture containing the DNA with two WT sites but no reb1 protein; D, ligation kinetics showing that both the substrate containing either two wt sites or a wt site and a M7 site yield approximately equal amounts of enhancement of ligation rate; E, measurement of ligation kinetics showing that a DNA substrate with one WT site and a M7 site show significant enhancement of ligation rate when incubated with the WT Reb1 but not with the mutant monomeric form of Reb1 protein, control experiments using the monomeric protein and DNA substrates with either two or a single WT Ter site did not show more than background levels of ligation enhancement; F, ligation kinetics showing that placing the WT and M7 sites in a mutually parallel orientation does not affect DNA looping.