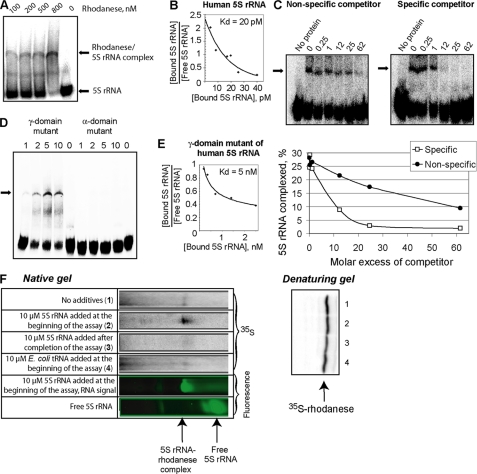
FIGURE 3.
Rhodanese-5 S rRNA interaction. A, mobility shift assay of complexes formed by human 5 S rRNA with urea-denatured rhodanese. The major rhodanese-5 S rRNA complex is marked with an arrow. The additional band may correspond to a minor complex (which was not always observed). B, Scatchard plot estimation of the dissociation constant for the rhodanese-5 S rRNA complex. C, mobility shift assays of rhodanese-5 S rRNA complexes formed in the presence of either nonspecific (yeast tRNA) or specific (nonlabeled human 5 S rRNA) competitors. Molar excess of competitors is marked above the autoradiographies. D, mobility shift assays of misfolded rhodanese complexed with a γ-domain (Δ(78–98)) or an α-domain (7G→U, 8G→A, and 9C→U) mutants of human 5 S rRNA. The total concentrations of rhodanese (in μm) are provided above. The major complex is marked with an arrow. E, Scatchard plot estimation of the dissociation constant for the complex of rhodanese with the γ-domain mutant of 5 S rRNA (Δ(78–98)). F, cotranslational binding of rhodanese to 5 S rRNA. The native gel (upper panel) shows the rhodanese-5 S rRNA complex formed during translation of rhodanese mRNAs in a rabbit reticulocyte system under various conditions. Uncomplexed rhodanese molecules remained on the top of the gel (data not shown). Lower panel, autoradiography of the same amounts of rabbit reticulocyte lysates separated by SDS-PAGE, showing the total amount of newly synthesized rhodanese.