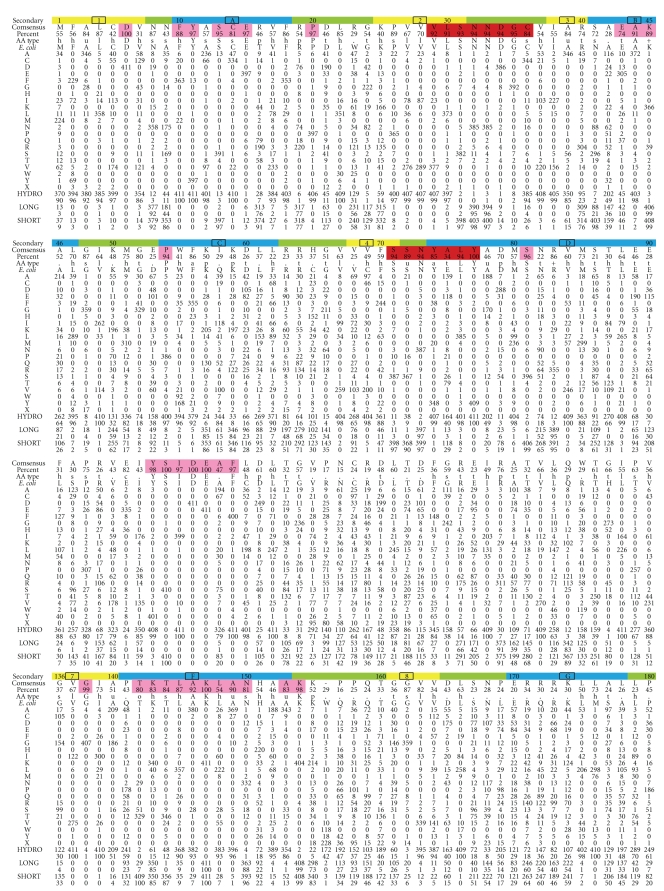
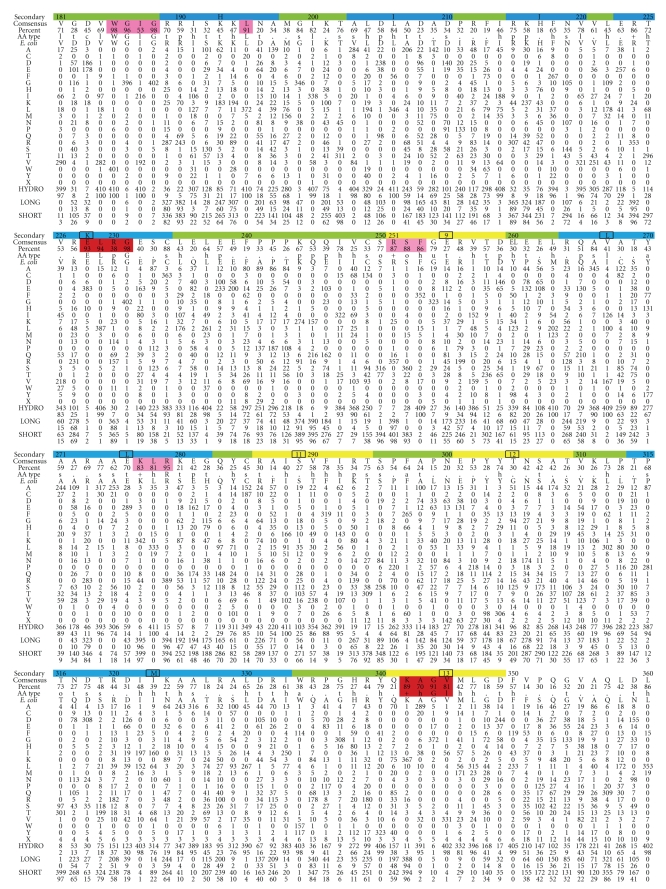
Figure 2.
Preferred amino acid at the 408 amino acid positions in UmuC(V), which is the polymerase subunit of DNAP V. Top row: amino acid numbering (every tenth residue) and the likely secondary structure based on alignment to other Y-Family DNAPs (β-strand, yellow; α-helix, blue, turn, green). Second row: consensus amino acid. Third row: percentage of the consensus amino acid. Fourth row: consensus type of amino acid, as codified in [101] (Website: http://coot.embl.de/Alignment/consensus.html). Regions where the consensus is high are highlighted in pink (all Y-Family DNAPs) or red (UmuC(V) only). Fifth Row: UmuC(V) sequence from E. coli. Rows 6–25 show the number of amino acids of each type at each position using the conventional one-letter code. Row 26: We note that some UmuC(V) sequences have slightly more or fewer than 422 amino acids. The number of UmuCs with an amino acid missing at a position compared to E. coli is shown in Row 26 (“X”). When an amino acid is present in a UmuC(V) but absent in E. coli, then the sequence is merely not included in Figure 2 (see text). In terms of gaps, E. coli only has one position that does not conform to the majority of UmuC(V)s: E. coli has an amino acid at position 156 (W), which is not present in a slight majority of all UmuC(V)s (226/408). The 408 UmuC(V) sequences were taken (as of 6/21/10) from the databases UniProt/Trembl [102] and HAMAP [103]. The sequences were aligned using MUSCLE [104]. Sequences were excluded for three reasons: (1) they were incomplete (i.e., sequences without recognizable versions of the N-terminus and the C-terminus were excluded); (2) they lacked any of the catalytic residues D6, D101, or E102; (3) they were redundant within a species (i.e., within a species if two UmuCs had the identical sequence, then only one was included).