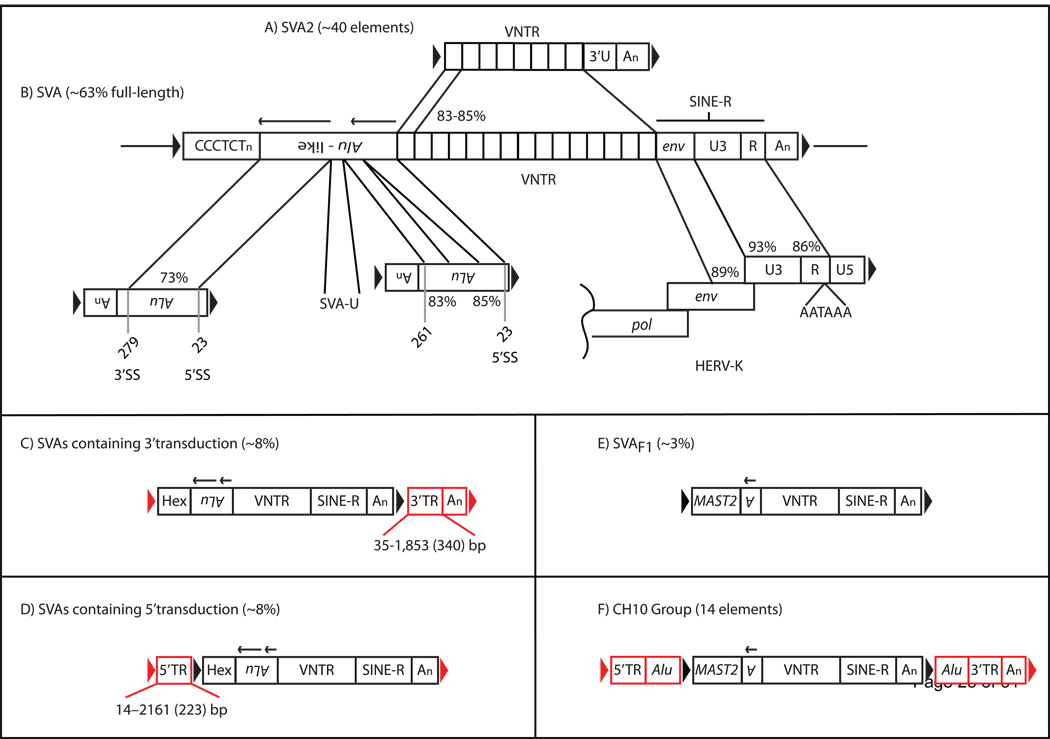
Figure 1.
The structure of a full-length SINE VNTR Alu (SVA) and SVA genomic variants. A) The SVA2 element. An SVA2 element consisting of a variable number of tandem GC-rich repeats (VNTR), followed by a unique 3’ sequence (3’U), followed by a polyA tail with the entire insertion flanked by a target-site duplication (black arrows) is shown. (B) A full-length SVA element consisting of in order from the 5’ end 1) CCCTCT hexameric repeats, 2) the Alu-like domain consisting of two antisense Alu fragments (black arrows above the Alu-domain indicate directionality of Alu sequences) and an intervening unique sequence, SVA-U, 3) a VNTR domain derived from the ancestral SVA2 element (A), 4) the SINE-R domain consisting of sequence sharing homology to the 3’end of the HERV-K10 env gene and U3, R, polyA signal (right LTR), terminating with a polyA tail (An) with the entire SVA insertion flanked by a target-site duplication. DNA sequence identities were obtained by pairwise BLAST alignments between the individual SVA domains and ancestral repeats (Alu, SINE-R, VNTR identity is between individual tandem repeats). Alignments consisted of using the following Repbase [27, 28] reference sequences: SVA2, SVA, Alu, HERV-K, and LTR5. The numbers below the antisense Alus correspond to nucleotide positions in AluRep and whether or not this position corresponds to a known splice site within Alu. Different SVA variants exist within the human genome, some contain additional 3’ sequence (C, red boxes), referred to as 3’ transductions (3’TR) or additional 5’ sequence (D, red boxes), referred to as 5’ transductions. A new target-site duplication (red arrows) flanks the SVA insertion and transduction. Transductions (C,D, red boxes) can be used to identify the source locus of a retrotransposon. Some 5’ transductions may be acquired via splicing of an upstream sequence into a downstream SVA element. This may result in novel SVA subfamilies such as SVAF1 (E) that acquired the MAST2 exon through splicing. SVA elements may also contain both 5’ and 3’ transductions (F), such as elements within the CH10 group. The number or percentage of SVAs containing a defining characteristic within the human genome reference sequence is in parentheses. Furthermore, the upper and lower limits in basepairs for reported 5’ [29] and 3’ [19] transductions is displayed below the transduction with the mean in parentheses.