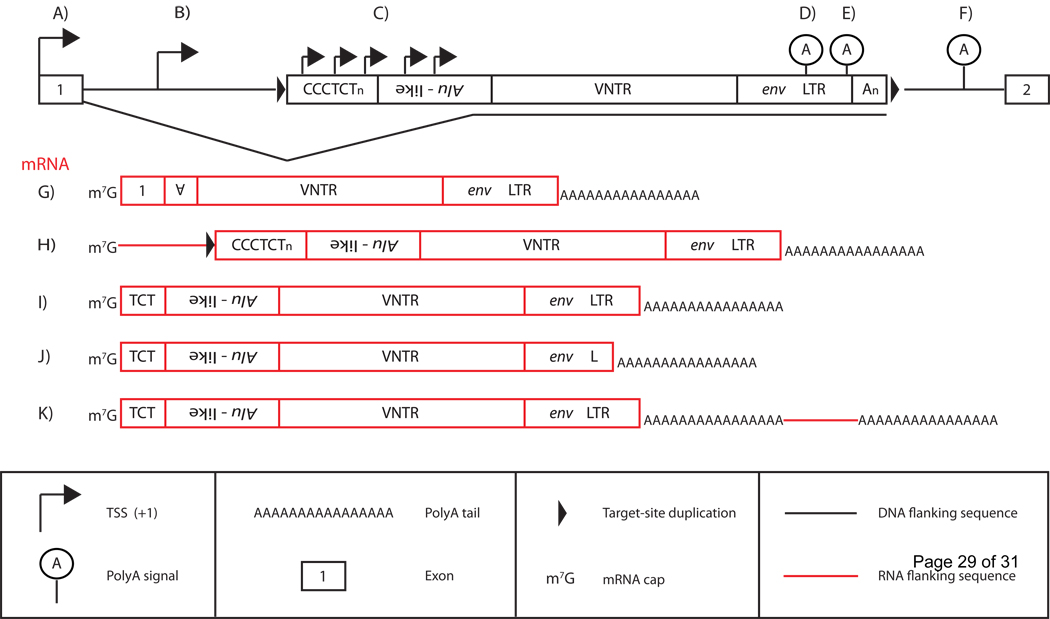
Figure 2.
SVA transcription and SVA mRNA structure. A full-length SVA, with individual domains labeled, in the genome residing in an intron (top; black line) of a gene with exons numbered 1 and 2 (top; black boxes) is displayed. Black bent arrowheads indicate different sites of SVA transcriptional initiation (A–C). The different SVA RNAs present in the human and chimpanzee transcriptome (red boxes; G–K) due to variable 5’ TSSs and 3’ polyA sites (D–F) are shown below. A) mRNA transcription may initiate at an upstream exon (black box labeled 1) that may subsequently splice into an SVA generating a “5’ truncated” SVA mRNA containing exonic sequence (G) terminating at the canonical SVA polyA signal (E). SVA-mediated exontrapping may enable SVA evolution as is the case for the SVAF1 subfamily. SVA transcription may initiate at an upstream TSS (B), presumably mediated by upstream promoter elements generating SVA mRNAs containing SVA 5’-flanking sequence (H; red line). Both (A,G) and (B,H) may result in retrotransposition of SVA 5’-flanking sequence, a process termed 5’-transduction. SVA RNAs may also initiate transcription internally (C) resulting in mRNAs resembling full-length genomic insertions (I), or terminate at internal non-canonical polyA sites in the SINE-R (D) resulting in 3’truncated SVA RNAs, or bypass the SVA polyA signal, terminating at a downstream polyA signal (F), resulting in an SVA mRNA containing 3’-flanking sequence (K; red line). Note that an SVA transcript may contain both 5’ and 3’ transductions (not shown).