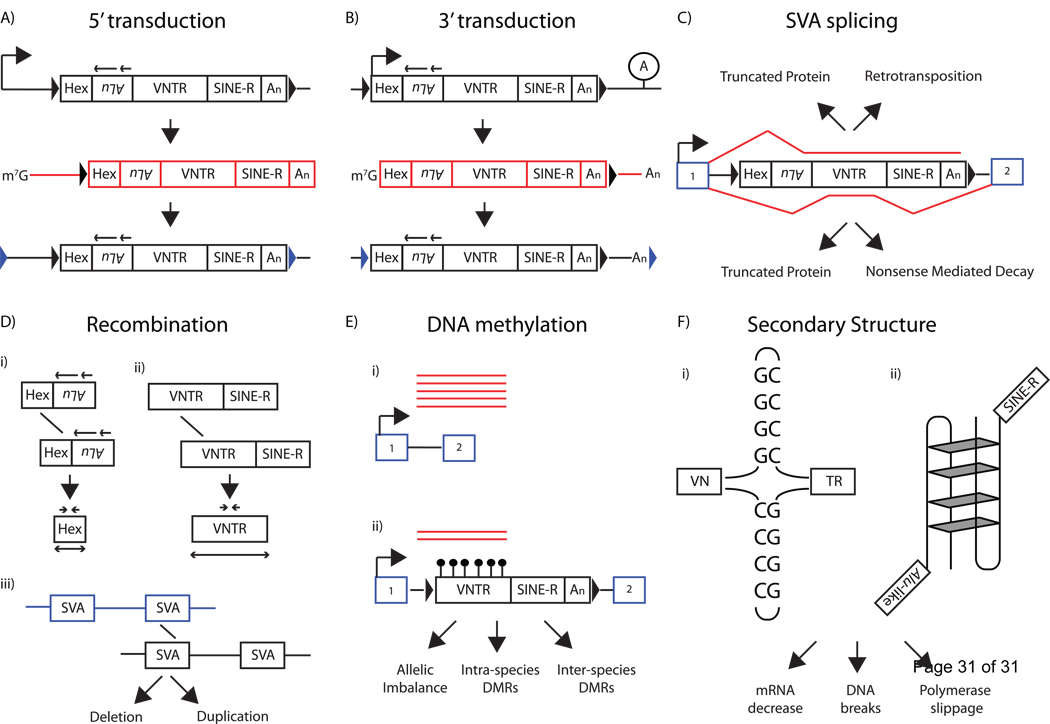
Figure 4.
Genomic Impact of SVA. A) SVAs are known to contain upstream transcriptional start sites (top, black bent arrow). This upstream transcriptional initiation will lead to an SVA mRNA (middle, red boxes) containing 5’ flanking sequence and sometimes result in retrotransposition of this sequence to a new genomic location (bottom), a process termed 5’ transduction. 5’ transduced sequence can be identified by the location of the target-site duplication positioned (blue arrowheads). B) SVAs are known to bypass their polyA signal and terminate mRNA transcription at a downstream polyA signal (lollipop A). This transcriptional readthrough will result in an SVA mRNA (middle, red boxes) containing 3’ flanking sequence and sometimes result in retrotransposition of this sequence to a new genomic location, a process termed 3’ transduction. 3’ transduced sequence can be identified by the location of the target-site duplication (blue arrowheads). The same SVA element is able to transduce 5’ and 3’ flanking sequences (not shown). C) SVA alternative splicing reduces host gene expression. SVA splicing may result in exon trapping (top, red line) that may result in truncated proteins or retrotransposition of upstream exons. SVA exonization (bottom, red line) may introduce nonsense codons into the mRNA leading to truncated proteins or nonsense-mediated decay. D) SVA elements may result in non-allelic homologous recombination within the CCCTCT hexamer (i) or the VNTR (ii), or (iii) between different SVA elements leading to deletion of genomic DNA. E) SVA VNTRs are known to be densely methylated (bottom, black lollipops) which may result in reduced mRNA expression (red lines) leading to allelic imbalance and inter- and intra- species-specific differentially methylated regions (DMR). F) The SVA VNTR is composed of GC-rich tandem repeats. Imperfect palindromes within individual tandem repeats may lead to the formation of cruciform structures in DNA (i). The VNTR may also result in more complex DNA structure, such as G-quadruplexes (ii). Structures formed due to the GC-richness of the VNTR may lead to decreases in mRNA, DNA breaks during replication, and DNA polymerase slippage, resulting in VNTR deletions. However, the RNA secondary structure of the VNTR is presumed to be functionally important in SVA retrotransposition.