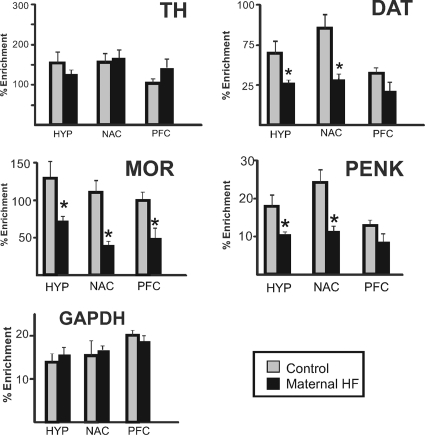
Figure 5.
DNA methylation changes at regulatory regions of reward-associated genes in offspring from HF-fed dams. Genomic DNA was isolated, sheared, and immunoprecipitated with 5-methylcytosine-specific antibody from brain regions from offspring of control (gray bars) or HF-fed (black bars) dams. The enrichment of DNA methylation relative to input genomic DNA at proximal promoter regions of TH, DAT, PENK, μ-opioid receptor (MOR), and GAPDH was quantified by qRT-PCR. Significant hypomethylation was observed in DAT, MOR, and PENK in offspring from HF-fed dams, which paralleled the increased expression of these genes. Values are mean ± sem. *, P values < 0.05 (n = 6 per group, two-tailed t test).