Table 1.
Thermal and Thermodynamic Stabilities of Triple Helical Complexes of Cross-Linked and Unlinked Pyrimidine Strands Binding Purine Sequencesa,b,d
| crosslink geometry | complex | pH = 7.0 | pH = 5.0 | ||
|---|---|---|---|---|---|
| Tm(°C) | -ΔG°37 (kcal/mol) | Tm(°C) | -ΔG°37 (kcal/mol) | ||
| no link (13mer) | 5′CTTCTTTTTCTTC | ||||
| 5′GAAGAAAAAGAAG | 13.3, 46.2c | -- | 48.0 | -- | |
| 3′CTTCTTTTTCTTC | |||||
| no link (14mer) | 5′CTTCTTTTTTCTTC | ||||
| 5′GAAGAAAAAAGAAG | 16.2, 47.9c | -- | 49.4 | -- | |
| 3′CTTCTTTTTTCTTC | |||||
| +0 link (13mer) | 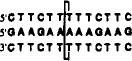 |
-- no crosslinking observed -- | |||
| +1 link (14mer) |  |
51.9 | 16.1 | 65.9 | 25.1 |
| +2 link (13mer) |  |
52.8 | 16.2 | 67.1 | 24.7 |
| +3 link (14mer) | 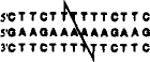 |
50.5 | 14.9 | 63.9 | 23.4 |
| +4 link (13mer) | 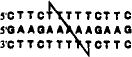 |
-- no crosslinking observed -- | |||
Conditions: 1.5 μM concentration each strand, 100 mM NaCl, 10 mM MgCl2, 10 mM Na·PIPES buffer.
Error limits for individual measurements are estimated at ±0.5 °C in Tm and ±5–10% in free energy.
First temperature is for triplex–duplex transition; second number is for duplex–single strands transition.
The positions of cross-linking groups are as indicated.
