Abstract
Defining the mechanisms governing myogenesis has advanced in recent years. Skeletal-muscle differentiation is a multi-step process controlled spatially and temporally by various factors at the transcription level. To explore those factors involved in myogenesis, stable isotope labeling with amino acids in cell culture (SILAC), coupled with high accuracy mass spectrometry (LTQ-Orbitrap), was applied successfully. Rat L6 cell line is an excellent model system for studying muslce myogenesis in vitro. When mononucleate L6 myoblast cells reach confluent in culture plate, they could transform into multinucleate myotubes by serum starvation. By comparing protein expression of L6 myoblasts and terminally differentiated multinucleated myotubes, 1170 proteins were quantified and 379 proteins changed significantly in fully differentiated myotubes in contrast to myoblasts. These differentially expressed proteins are mainly involved in inter-or intracellular signaling, protein synthesis and degradation, protein folding, cell adhesion and extracelluar matrix, cell structure and motility, metabolism, substance transportation, etc. These findings were supported by many previous studies on myogenic differentiation, of which many up-regulated proteins were found to be involved in promoting skeletal muscle differentiation for the first time in our study. In sum, our results provide new clues for understanding the mechanism of myogenesis.
Keywords: Quantitative proteomics, SILAC, Skeletal-muscle differentiation, 2D-LC-LTQ-Orbitrap
1 Introduction
Skeletal-muscle differentiation is a complicated process coordinated by several transcription factors [1, 2]. Under the control of those transcription factors, proliferating myoblasts withdraw from the cell cycle, and then elongate, adhere, and fuse into multinucleated myotubes. Finally, matured myotubes convert into myofibres, which are capable of muscle contraction. A number of muscle differentiation factors have been discovered such as the myogenic regulatory factors (MRFs) and the myocyte enhancer binding-factors (MEFs). The expression of these transcription factors, such as MyoD, Myogenin, Myf5 and Mef2, are controlled positively by the P38/MAPK, Wnt and Sonic hedgehog (Shh) pathways, and are inhibited by BMPs and the Notch/Delta pathway in muscle precursors [1, 3]. When the positive regulation factors are dominant, the transcriptions of muscle-specific genes are activated and the differentiation process is initiated. Although the main factors orchestrating skeletal-muscle differentiation are well defined, little is known about how these growth factors and signal pathways act on myogenic differentiation synergistically [1–3]. When myoblasts proliferate to skeletal cells, many characteristics, from the morphological to the conformational, will change significantly. It is reasonable to speculate that many additional cellular components are involved in myogenesis. Accumulating evidences suggest that myogenesis was regulated spatio-temporally by many cellular components; therefore, identifying additional components underlying networks that promote skeletal-muscle differentiation could lead to new insights into the process of myogenesis.
Quantitative proteomics allows measurement of differential protein expression [4, 5]. Tannu et al examined total cellular proteins, membrane-, and nuclear-enriched proteins using 2-D gel electrophoresis between proliferating mouse myoblasts of C2C12 cells and fully differentiated myotubes. [6]. The proteins they identified are mainly involved in cell signaling, cell cycle and cell shape in differentiating C2C12 cells. Gonnet and colleagues identified 105 proteins with expressional variance in differentiating human myoblasts of different myogenic period by 2D DAGE. They found that some unique proteins may participate in human muscle differentiation [7]. Kislinger et al used a gel-free shotgun proteomics method together with label-free quantitative proteomics to profile expression changes in crude nuclei during differentiation stages [8]. Hierarchical clustering of the resulting protein profiles and gene expression found that several types of proteins may be involved in myogenic process, such as integrin, septin. On the whole, these studies have presented more information in principle about the characterization of skeletal muscle differentiation by proteomics methods, but hard work on myogenesis still need to be done because of the very complex myogenic process. Moreover, there are many divergences amongst the previous studies based on molecular methods or proteomics methods. To further discover additional information about the differentiation process, we sought to use stable isotope labeling methods together with shotgun proteomics to quantitate protein expression. Stable isotope labeling with amino acids in cell culture (SILAC) has been combined with highly sensitive tandem mass spectrometry to create a simple, straightforward, and efficient approach for large-scale protein quantification [5,9,10]. SILAC relies on metabolic incorporation of a “light” or “heavy” isotopic form of the amino acid into cellular proteins [9]. SILAC have been applied in various biological fields to detect the biological changes of protein abundance, protein modifications states, and protein-protein interactions [10]. Ong and colleagues used the myogenic differentiation of C2C12 cells as a model to establish and confirm the SILAC method, but they didn’t present the myogenesis-related proteins in details [9]. In this study, we employed SILAC method with 2D-LC and LTQ-Orbitrap Hybrid Mass Spectrometer to determine protein expression differences between rat L6 myoblasts and myotubes for the first time.
2 Materials and Methods
Materials
Analytical grade chemicals were obtained from Sigma (St. Louis, USA). Milli-Q water was used unless otherwise mentioned. Normal high glucose DMEM media, fetal bovine serum (FBS), glutamine, sodium pyruvate, PBS, penicillin and streptomycin were purchased from Invitrogen (Carlsbad, CA, USA). DMEM media deficient in arginine was purchased from JRH Biosciences (Lenexa, KA, USA). Dialyzed FBS was purchased from Biological Industries (Kibbutz Beit Haemek, Israel). Both Light 12C6 14N4 L- arginine and heavy 13C6 15N4 L-arginine were obtained from Spectra Stable Isotopes (Columbia, KS, USA). Protease inhibitor cocktail tablets were obtained from Roche (Basel, Switzerland). Sequence grade trypsin was purchased from Promega (Madison, WI, USA). HPLC grade acetonitrile, methanol, and formic acid were obtained from J. T. Baker (Phillipsburg, PA, USA). Primary antibodies to tubulin-β, MyoD, desmin, 14-3-3γ, Prohibitin-2, and HRP-conjugated secondary antibodies were purchased from Abcam (Cambridge, UK). HRP-conjugated Primary antibody to GADPH was purchased from Kangcheng (Shanghai, China). SuperSignal® west Femto trial kit was obtained from Pierce (Rockford, IS, USA).
Cell culture and isotopic metabolic labelling
Rat L6 myoblasts were maintained in DMEM with 4mM L-glutamine, 4.5g/L glucose, 50 UI/ml Penicillin and 50 ug/ml streptomycin, additionally supplemented with 10% (v/v) FBS (growth medium,GM). Once myoblasts reached confluence, differentiation was induced by lowering the serum concentration to 2% (differentiation medium, DM). For western blot, L6 myoblasts in common media were subcultured into six 100mm of culture plates. After differentiation was induced, media were changed every 48hrs. At day 0, day1, day2, day3, day4, and day8, one plate of cells was washed by cold PBS separately and kept at −80 °C for protein extraction later. For isotopic metabolic labeling, newly subcultured L6 cells were transfered into DMEM supplemented with 8% dialyzed FBS plus 2% normal FBS and light 12C6 14N4 L-arginine or heavy 13C6 15N4 L-arginine instead of common GM. L6 myoblasts in light media were induced into myotubes. L6 myoblasts were subcultured in heavy 13C6 15N4 L-arginine for at least seven population doublings. Light myotubes and heavy myoblasts were washed three times with ice-cold PBS separately for protein extraction.
Protein extraction
The process of protein extraction for either MS analysis or western blot is same. The following steps were carried out at 4 °C. Cells were scraped into 8M urea with protease inhibitor cocktail tablet (Roche, Basel, Switzerland) and sonicated for cells lysis separately. After centrifugation for 30 min at 20,000g in a bench-top centrifuge (Thermo Fisher Scientific, Waltham, MA, USA), the supernatants were collected and kept at −80°C for analysis. Protein concentrations were measured using the Bradford method.
In-solution digestion
Extracted protein samples from heavy myoblasts and light myotubes were combined at a 1:1 ratio.In-solution digestion was performed with the following protocol. Briefly, 100ug of protein mixture was dissolved in 8M urea and 25mM NH4HCO3, reduced with 10mM DTT for 1 hour, alkylated by 40mM iodacetamide in the dark for 45 minutes at room temperature, and then 40mM DTT was added to quench the iodacetamide for 30 min at room temperature. After diluting 8M urea with 25mM NH4HCO3 to 1.6 M, sequence grade trypsin was added at a ratio of 1:30 and digested at 37 °C for overnight. Tryptic digestion was stopped by adding formic acid to a 1% final concentration.
2D-LC-MS/MS analysis
Digests were centrifuged at 16000g for 10 min prior to analysis. The supernatant was analyzed by two dimensional liquid chromatography (2D-LC) on an LTQ Orbitrap XL (Thermo Fisher Scientific, Waltham, USA) following the method below [11]. For single analyses, 100 µg of peptide mixtures were pressure-loaded onto a two-dimensional silica capillary column packed with 3cm of C18 resin (Synergi 4u Hydro-RP 80A, Phenomenex, CA, USA) and 3cm of strong cation exchange resin (Luna 5u SCX 100A, Phenomenex, USA) . The buffer solutions used were 5% acetonitrile/0.1% formic acid (buffer A), 80% acetonitrile/0.1% formic acid (buffer B), and 500 mM ammonium acetate/5% acetonitrile/0.1% formic acid (buffer C). The two-dimensional column was first desalted with buffer A and then eluted using an eight-step salt gradient ranging from 0 to 500 mM ammonium acetate. The effluent of the two-phase column in each case was directed onto a 10cm of C18 analytical column (100 µm i.d.) with a 3–5 µm spray tip. Step 1 consisted of a 100-min gradient from 0%–100% buffer B. Steps 2–9 had the following profile: 3 min of 100% buffer A, 3 min of X% buffer C, a 10-min gradient from 0%–15% buffer B, and a 97-min gradient from 15%–55% buffer B. The 3-min buffer C percentages (X) were 5%, 10%, 15%, 20%, 30%, 40%, 55%, and 75% respectively, for the 8-step analysis. The final step, the gradient contained: 3 min of 100% buffer A, 20 min of 100% buffer C, a 10-min gradient from 0%–15% buffer B, and a 107-min gradient from 15%–70% buffer B. Nano-electrospray ionization was accomplished with a spray voltage of 2.5 kV and a heated capillary temperature of 230°C. A cycle of one full-scan mass spectrum (400–2000 m/z) followed by six data-dependent tandem mass spectra was repeated continuously throughout each step of the multidimensional separation. All tandem mass spectra were collected using normalized collision energy (a setting of 35%), an isolation window of 3 m/z, and 1 micro-scan. Application of mass spectrometer scan functions and HPLC solvent gradients were controlled by the XCalibur data system (Thermo Fisher, Waltham, USA).
Data analysis and bioinformatics
MS and tandem mass spectra were extracted from the XCalibur data system format (.RAW) into MS1 and MS2 formats (Mc-Donald et al. 2004) by RAW_Xtractor [12]. The target database was the EBI-IPI rat database; the target database was attached with common contaminants such as keratins; the whole database (target + contaminants) was then reversed and attached. Tandem mass spectra were interpreted by SEQUEST using an EBI-IPI rat database (Version 3.17, 2006). Sequences for common contaminants such as keratins, IgGs, protease autolysis products are added to the database and then a copy is reversed and appended. Results were filtered, sorted, and displayed using the DTASelect2 program [13]. Only peptides with ≥ 95% confidence score, maximum Sp rank of 1000 and a ΔCn score of ≥ 0.1 were considered. In addition, a minimum sequence length of seven amino acid residues was required. The false positive rate for protein identification was kept below 1%. Quantitative ratios were determined by the software CenSus version 0.9 [14]. The annotations of proteins were obtained from SwissProt and TrEMBL protein database. For proteins without descriptions, annotations was done by searching the IPI, SwissProt and TrEMBL protein database with BlastP for homologous proteins with descriptions. The PANTHER classification system was used for protein sorting (www.pantherdb.org) with slight modification where a few protein groups with similar annotation were combined. The STRING (http://string.embl.de/), a proteins and their interactions prediction system, was used to retrieve protein associations.
Western blotting
L6 cells were washed three times with cold PBS and protein was extracted as described above. Equivalent amounts of protein (10µg per lane) were separated by SDS-PAGE and electroblotted onto 0.45-um HybondTM-P PVDF membranes (GE healthcare, Piscataway, NJ, USA) by the semi-dry method. Binding of nonspecific proteins to membranes was blocked by incubating these in blocking buffer consisting of 5% non-fat milk in TBS plus 0.05% Tween 20 (TBST) for 1 h at 25°C. Membranes were then incubated at 4°C overnight with primary antibodies diluted in blocking buffer. Membranes were washed three times with TBST, incubated with HRP-conjugated secondary antibodies for 1 h, and then washed three times again with TBST. Finally, immunoreactive proteins on the membranes were detected by SuperSignal® west Femto trial kit and exposed to x-ray film. Western blots were scanned and gray scales were quantified by ImageQuant TL (GE healthcare, Piscataway, NJ, USA). L6 cells cultured for western blotting were harvested three times. Western blotting for every selected protein was repeated three times for every batch of total protein extract.
3 Result and discussion
3.1. Morphological conversion of L6 Cells
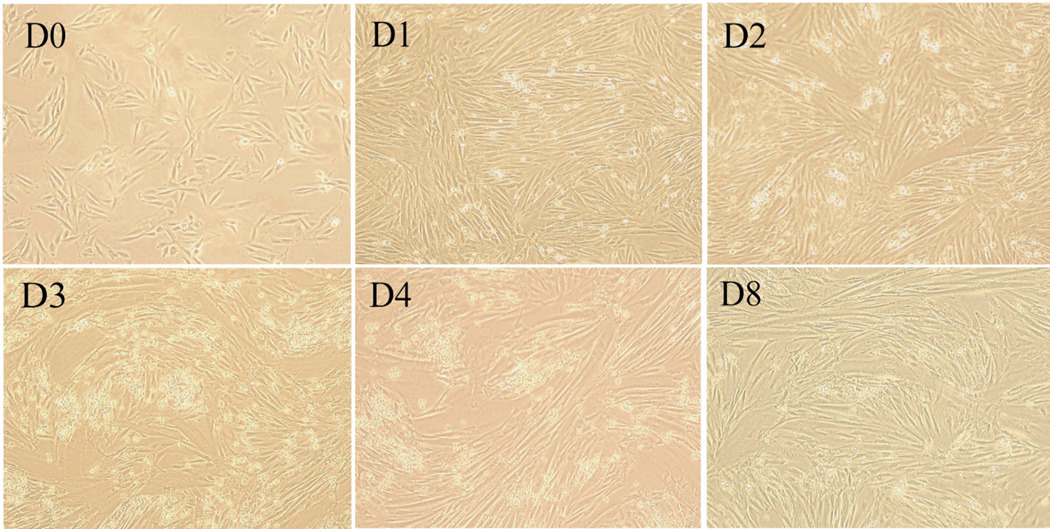
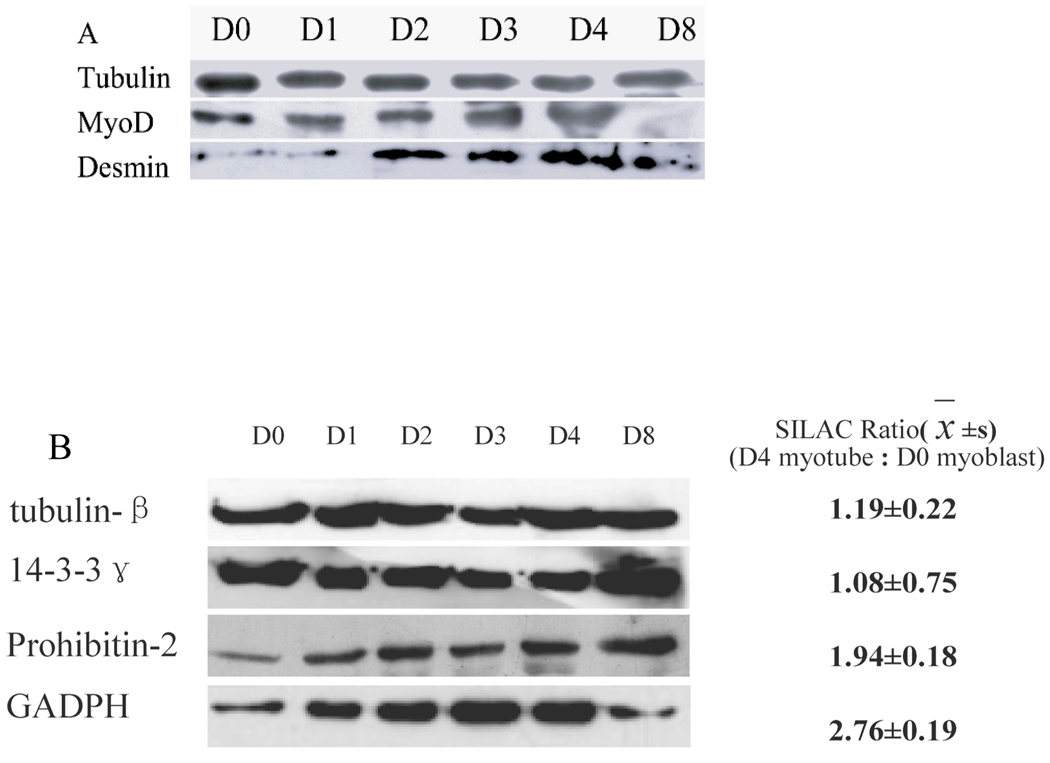
Rat L6 cell is an excellent model system for developmental biology associated with cell proliferation, signal transduction and cell fate determination. Normally, L6 myoblast cells were cultured in DMEM media supplemented with 10% fetal bovine serum. In our experiments, with fetal bovine serum decreasing from 10% to 2% in DMEM media, mononucleate L6 myoblast in good conditions fused and transformed into multinucleate myotubes very quickly. One day latter after serum deprivation, myocytes are scattered in the cell culture plate. In spite of that, the shapes of L6 cells were distinct from myoblasts. After 2 days, about 35% area of culture plate was occupied by myocytes. After 3 days more than 85% of cells have fused into elongated myotubes. At the end of day 4, giant elongated multinucleate myotubes have outspread apparently everywhere on the culture plate. As Fig. 1 shows, the morphology of cells from myoblasts to myotubes has changed significantly (Figure 1). Accompanied with the morphological conversion of myoblast into myotubes, some muscle-specific proteins were expressed in high level in myotubes in comparison with myoblasts. We examined muscle-differentiation marker desmin and MyoD by western blot (Figure 2a). As the figure 2a shows that the expressional levels of these proteins were increased gradually during myogenic process but decreased in D8 myotubes. In addtion, our SILAC results show that some other muscle-differentiation markers, such as myosin heavy chain (MHC), skeletal muscle actin alpha, and myosin heavy polypeptide 2, also were up-regulated during myogenic process. In conclusion, L6 myoblasts had well differentiated into myotubes.
Figure 1.
The light micrographs of cultured L6 cells were enlarged by 10*10. D0 represents that the myoblasts before differentiation initiation were grown in DMEM supplemented with 10% FBS. D1, D2, D3 and D4 indicates that L6 cells have been cultured in DMEM with 2% FBS for one, two, three and four days, respectively. D4 were fully differentiated myotubes. D8 indicates myotubes have been serum-starved for another four days starting from D4.
Figure 2.
Western blotting. (A) Western blotting of L6 differentiation marker or differentiation associated prtoteins in L6 blasts to L6 myotubes. (B) Validation of SILAC ratio by western blotting.
3.2. Protein identification and quantification
The morphological conversion of L6 cells from myoblasts to myotubes is presumed to be driven by proteins that follow tissue- and cell specific expression. The goal of this work was to find expression differences between rat L6 myoblasts and myotubes. The tryptic peptides were analyzed by 2D-LC-LTQ-Orbitrap MS system [11]. For protein identification, a decoy database was used and the false positive rate for protein identification was kept below 1% in this study. After filtering with stringent parameters, 12,729 peptides and 2,767 proteins were identified from all experiments (see supplementary table 1). Among these proteins, 1170 proteins were quantified with high confidence (see supplementary table 1 and 2). 780 proteins (about 67%) were quantified with two or more peptides (see supplementary figure 1). Among the quantified proteins, 342 proteins were up-regulated (≥1.5-fold changes, see Table1) and 37 proteins were down-regulated (≤0.5-fold changes) in fully differentiated myotubes (see Table 2). The left proteins were considered as no significant changes (see supplementary table 1). Our results show that the expression of many conserved proteins, such as tubulin beta chain, tubulin beta 2, tubulin β 5, and actin β (actin, cytoplasmic 1), remains stable, which suggests the accuracy of the quantitative results in our experiment to some extent. To further validate the accuracy of the quantitative results by different methods, several proteins with different SILAC quantitative ratio were quantified again by western blotting. Figure 3 shows the results of western blotting for tubulin-β, 14-3-3γ, prohibitin-2 and GADPH are strongly consistent with the quantitative results determined by mass spectrometry.
Table 1.
Classification of Proteins with the Ratio ≥1.5
| Description | IPI ID | Average Ratio Average±SD |
Quantification Peptide Num |
UniProtKB ID/AC |
Gene ID |
|---|---|---|---|---|---|
| Mediators of Signaling Pathway | |||||
| Dual specificity mitogen-activated protein kinase 1 (MAPKK 1) |
IPI00231247.8 | 1.75±NA | 1 | Q01986 | 170851 |
| Guanine nucleotide-binding protein G(i), alpha-2 subunit |
IPI00231925.7 | 1.79 ±0.01 | 3 | P04897 | 81664 |
| Hypothetical protein LOC361120 | IPI00364617.2 | 1.53±NA | 1 | Q4QQT7 | 361120 |
| Inositol 1,4,5-trisphosphate receptor type 3(IP3R-3) | IPI00206986.1 | 1.55±NA | 1 | Q63269 | 25679 |
| latent transforming growth factor beta binding protein 1 |
IPI00389331.1 | 1.80±0.00 | 2 | Q00918 | 59107 |
| Membrane associated progesterone receptor component 2 |
IPI00373197.1 | 2.15±0.15 | 2 | Q5XIU9 | 361940 |
| Phosphatidylethanolamine-binding protein | IPI00230937.4 | 1.62±NA | 1 | P31044 | 29542 |
| PREDICTED: A kinase (PRKA) anchor protein 2 | IPI00364858.3 | 4.05±NA | 1 | Q5U301 | 298024 |
| PREDICTED: similar to Dual specificity protein phosphatase 3 |
IPI00568511.1 | 1.95±NA | 1 | 498003 | |
| PREDICTED: similar to mitogen-activated protein kinase 4 isoform (Map4k4) |
IPI00357885.3 | 2.28±NA | 1 | 301363 | |
| PREDICTED: similar to Ran-binding protein 10 | IPI00365184.2 | 1.87±NA | 1 | 361396 | |
| PREDICTED: similar to receptor expression enhancing protein 5 |
IPI00360246.2 | 3.34 ±0.22 | 2 | 364838 | |
| PREDICTED: similar to Semaphorin 3C | IPI00372650.3 | 1.88±NA | 1 | 296787 | |
| Prenylated Rab acceptor protein 1 | IPI00208565.1 | 2.18±NA | 1 | O35394 | 83583 |
| Prohibitin-2 | IPI00190557.2 | 1.94 ±0.18 | 4 | Q5XIH7 | 114766 |
| RAB10, member RAS oncogene family | IPI00555185.1 | 2.18±0.02 | 3 | Q5RKJ9 | 50993 |
| Ras-related protein Rab-2A | IPI00202570.1 | 1.50±NA | 1 | P05712 | 65158 |
| Reticulocalbin 3, EF-hand calcium binding domain | IPI00207050.5 | 55.97±NA | 1 | Q63399 | 494125 |
| SH2-containing inositol phosphatase 2 | IPI00205291.1 | 3.02±1.10 | 2 | Q9WVR3 | 65038 |
| Signal sequence receptor, alpha | IPI00561555.1 | 1.78±NA | 1 | Q4V7D1 | 361233 |
| Splice Isoform 3 of Protein kinase C and casein kinase substrate in neurons 2 protein |
IPI00231216.1 | 1.55±NA | 1 | Q9QY17 | 124461 |
| Striatin, calmodulin binding protein 3 | IPI00476338.2 | 2.37±0.19 | 4 | P58405 | 114520 |
| Translocon-associated protein gamma subunit | IPI00196589.2 | 1.67±NA | 1 | Q08013 | 81784 |
| A-kinase anchor protein 11 | IPI00210194.1 | 1.84±NA | 1 | Q62924 | 498549 |
| Protein Synthesis | |||||
| 3-hydroxyisobutyrate dehydrogenase, mitochondrial precursor |
IPI00202658.1 | 2.05±NA | 1 | P29266 | 63938 |
| 60S ribosomal protein L5 | IPI00230914.4 | 3.83 ±3.73 | 5 | P09895 | 81763 |
| Asparaginyl-tRNA synthetase | IPI00421313.1 | 1.99 ±0.27 | 7 | Q6TUD1 | 291556 |
| Cc2-5 | IPI00382192.1 | 4.16±NA | 1 | Q7TP13 | 307759 |
| Dolichyl-diphosphooligosaccharide--protein glycosyltransferase 63 kDa subunit precursor |
IPI00188059.2 | 1.65 ±0.05 | 3 | P25235 | 64701 |
| Elongation factor 1-alpha 1 | IPI00195372.1 | 1.58 ±0.04 | 13 | P62630 | 171361 |
| Elongation factor 2 | IPI00203214.5 | 1.61 ±0.10 | 18 | P05197 | 29565 |
| ERGIC-53 protein precursor | IPI00210116.2 | 1.54 ±0.06 | 3 | 116666 | |
| Eukaryotic translation elongation factor 1 delta | IPI00471525.2 | 2.07 ±0.07 | 8 | Q68FR9 | 300033 |
| Eukaryotic translation initiation factor 2 subunit 1 | IPI00230830.4 | 1.73±NA | 1 | P68101 | 54318 |
| Eukaryotic translation initiation factor 3 subunit 9 | IPI00363771.3 | 1.50 ±0.06 | 6 | Q4G061 | 288516 |
| Eukaryotic translation initiation factor 4A2 | IPI00193595.3 | 2.07 ±0.96 | 7 | Q52KC1(m) | 303831 |
| GERp95 | IPI00214529.3 | 1.91±NA | 1 | Q9QZ81 | 59117 |
| Glutamyl-prolyl-tRNA synthetase | IPI00421357.2 | 1.98 ±0.32 | 3 | Q6TXE9 | 289352 |
| Hypothetical protein LOC619440 | IPI00201819.1 | 1.64 ±0.05 | 2 | Q5XI97 | 619440 |
| IKIP1 protein | IPI00199778.1 | 1.62±0.05 | 3 | Q5EAJ6 | 314730 |
| Leucine rich repeat containing 47 (predicted) | IPI00359172.1 | 2.00 ±0.00 | 2 | 362672 | |
| Leucyl-tRNA synthetase | IPI00363236.2 | 1.57±NA | 1 | Q5PPJ6 | 291624 |
| Phenylalanine-tRNA synthetase-like, beta subunit | IPI00202379.1 | 1.71 ±0.31 | 3 | Q68FT7 | 301544 |
| PREDICTED: eukaryotic translation elongation factor 1 gamma |
IPI00470317.3 | 3.04 ±0.03 | 2 | Q68FR6 | 293725 |
| PREDICTED: similar to Eif3s1 protein | IPI00364189.1 | 1.50 ±0.10 | 2 | A0JPM9 | 311371 |
| PREDICTED: similar to Eukaryotic translation elongation factor 1 beta 2 |
IPI00476899.1 | 2.45±NA | 1 | 363241 | |
| PREDICTED: similar to eukaryotic translation initiation factor 3, subunit 10 theta, 150/170kDa |
IPI00372810.2 | 1.59 ±0.11 | 2 | Q4G020 | 292148 |
| PREDICTED: similar to Glycyl-tRNA synthetase | IPI00364262.2 | 1.90 ±0.06 | 3 | Q5I0G4 | 297113 |
| PREDICTED: similar to isoleucine-tRNA synthetase | IPI00365783.3 | 2.04±NA | 1 | Q6NXK4(m) | 306804 |
| PREDICTED: similar to Methionine-tRNA synthetase |
IPI00366397.2 | 1.71±0.00 | 2 | 299851 | |
| PREDICTED: similar to Mitochondrial 28S ribosomal protein S30 |
IPI00214903.3 | 1.66±NA | 1 | Q9D0G0(m) | 294767 |
| PREDICTED: similar to Signal recognition particle 68 |
IPI00368134.2 | 1.63 ±0.24 | 2 | 363707 | |
| PREDICTED: similar to Tryptophanyl-tRNA synthetase |
IPI00365914.2 | 2.40 ±0.02 | 3 | Q6P7B0 | 314442 |
| PREDICTED: similar to Tu translation elongation factor, mitochondrial |
IPI00371236.2 | 1.56 ±0.00 | 2 | 293481 | |
| Probable ATP-dependent RNA helicase DDX46 | IPI00208266.2 | 1.59±NA | 1 | Q62780 | 245957 |
| Prolyl 4-hydroxylase alpha-1 subunit precursor | IPI00209863.2 | 1.97 ±0.26 | 2 | P54001 | 64475 |
| Prolyl 4-hydroxylase alpha-2 subunit precursor | IPI00372370.2 | 1.77±0.00 | 2 | 360526 | |
| Ribophorin I | IPI00204365.2 | 1.63 ±0.10 | 5 | Q6P7A7 | 25596 |
| Seryl-aminoacyl-tRNA synthetase 1 | IPI00373410.3 | 2.40 ±0.09 | 4 | Q6P799 | 266975 |
| syntaxin 12 | IPI00208759.2 | 1.74±NA | 1 | O88385 | 65033 |
| Threonyl-tRNA synthetase, cytoplasmic | IPI00559880.1 | 1.98±0.04 | 3 | Q5XHY5 | 294810 |
| Transcriptional activator protein Pur-beta | IPI00189358.2 | 3.76 ±0.80 | 3 | Q68A21 | 498407 |
| Transitional endoplasmic reticulum ATPase | IPI00212014.2 | 1.50±0.13 | 13 | P46462 | 116643 |
| Tyrosyl-tRNA synthetase | IPI00366785.2 | 1.66±NA | 1 | Q4KM49 | 313047 |
| WD repeat domain 77 | IPI00368916.1 | 1.59±NA | 1 | Q4QR85 | 310769 |
| Proteolysis | |||||
| 26S protease regulatory subunit 4 | IPI00211733.1 | 1.88±NA | 1 | P62193 | 117263 |
| 26S protease regulatory subunit 7 | IPI00421600.7 | 1.78 ±0.10 | 4 | Q63347 | 25581 |
| ATPase family AAA domain-containing protein 1 | IPI00566676.1 | 1.55±NA | 1 | 309532 | |
| Dipeptidyl-peptidase 2 precursor | IPI00230946.4 | 3.14±NA | 1 | Q9EPB1 | 83799 |
| Fxna | IPI00390597.2 | 1.73±NA | 1 | Q6UPR8 | 373544 |
| Insulin-like growth factor binding protein 5 protease | IPI00199325.1 | 4.11 ±0.24 | 4 | Q9QZK5 | 65164 |
| Lon | IPI00205076.1 | 2.19 ±0.01 | 3 | Q924S5 | 170916 |
| Midline-1 | IPI00231578.4 | 2.65±NA | 1 | P82458 | 54252 |
| Mitochondrial-processing peptidase beta subunit, mitochondrial precursor |
IPI00209980.5 | 2.35±NA | 1 | Q03346 | 64198 |
| PREDICTED: aminopeptidase puromycin sensitive | IPI00372700.1 | 2.22 ±0.15 | 5 | Q8VID2 | 50558 |
| PREDICTED: proteasome (prosome, macropain) subunit, beta type 5 |
IPI00230992.4 | 2.50±NA | 1 | Q91X53 | 29425 |
| PREDICTED: similar to 26S proteasome non-ATPase regulatory subunit 11 |
IPI00370382.1 | 1.52 ±0.03 | 3 | Q8BG32 | 303353 |
| PREDICTED: similar to 26S proteasome subunit p40.5 |
IPI00202283.1 | 1.58 ±0.49 | 2 | Q9WVJ2 | 365388 |
| PREDICTED: similar to Expressed sequence AI314180 |
IPI00367234.2 | 1.76 ±0.08 | 3 | 313196 | |
| PREDICTED: similar to HECT domain containing 1 | IPI00365611.2 | 1.97±NA | 1 | 362736 | |
| PREDICTED: similar to Psmc6 protein | IPI00362105.1 | 1.82±NA | 1 | Q32PW9 | 289990 |
| Prenylcysteine oxidase precursor | IPI00198080.1 | 1.54±NA | 1 | Q99ML5 | 246302 |
| Proteasome (Prosome, macropain) 26S subunit, ATPase 3 |
IPI00190392.3 | 1.52±NA | 1 | Q63569 | 29677 |
| Proteasome (Prosome, macropain) 26S subunit, non-ATPase, 2 (PSMD2) |
IPI00370456.1 | 1.58 ±0.19 | 10 | Q4FZT9 | 287984 |
| Proteasome 26S subunit, non-ATPase, 3 | IPI00370009.1 | 1.56 ±0.05 | 5 | Q5U2S7 | 287670 |
| Proteasome subunit alpha type 1(PSMA1) | IPI00191748.3 | 1.72±NA | 1 | P18420 | 29668 |
| Proteasome subunit alpha type 2 | IPI00231757.11 | 1.77 ±0.07 | 3 | P17220 | 29669 |
| Proteasome subunit alpha type 3 | IPI00476178.2 | 4.42±0.01 | 2 | Q6IE67 | 408248 |
| Proteasome subunit alpha type 4 | IPI00231046.8 | 1.61 ±0.00 | 2 | P21670 | 29671 |
| Proteasome subunit alpha type 5 | IPI00191502.5 | 1.50±0.03 | 5 | P34064 | 29672 |
| Proteasome subunit beta type 1 (PSMB1) | IPI00191749.5 | 1.64 ±0.28 | 3 | P18421 | 94198 |
| Proteasome subunit beta type 2 | IPI00188584.1 | 1.61 ±0.02 | 3 | P40307 | 29675 |
| Proteasome subunit beta type 4 precursor | IPI00191505.3 | 1.60±NA | 1 | P34067 | 58854 |
| Proteasome, 26S, non-ATPase regulatory subunit 6 | IPI00189463.1 | 1.70 ±0.05 | 4 | Q6PCT9 | 289924 |
| Protective protein for beta-galactosidase | IPI00464785.1 | 1.91 ±0.00 | 2 | Q6AYS3 | 296370 |
| Prothrombin precursor (Fragment) | IPI00189981.1 | 5.18±NA | 1 | P18292 | 29251 |
| Secernin 1 | IPI00202627.1 | 1.83±NA | 1 | Q6AY84 | 502776 |
| Tripeptidyl-peptidase 2 | IPI00213579.2 | 1.59±NA | 1 | Q64560 | 81815 |
| Ubiquitin-conjugating enzyme E2 variant 2 | IPI00339040.2 | 1.67 ±0.12 | 2 | Q7M767 | 287927 |
| Molecular Chaperone | |||||
| 10 kDa heat shock protein, mitochondrial | IPI00326433.10 | 2.20 ±0.04 | 3 | P26772 | 25462 |
| 60 kDa heat shock protein, mitochondrial precursor | IPI00339148.2 | 1.53 ±0.04 | 8 | P63039 | 63868 |
| 78 kDa glucose-regulated protein precursor | IPI00206624.1 | 1.75 ±0.06 | 13 | P06761 | 25617 |
| Calnexin Precursor | IPI00199636.1 | 1.76±0.23 | 8 | P35565 | 29144 |
| Chaperonin subunit 6a | IPI00188111.1 | 1.50±0.06 | 6 | Q3MHS9 | 288620 |
| Heat shock 70 kDa protein 4/Ischemia responsive 94 kDa protein |
IPI00387868.2 | 1.83±0.12 | 3 | O88600 | 266759 |
| Heat shock cognate 71 kDa protein | IPI00208205.1 | 1.58 ±0.07 | 8 | P63018 | 24468 |
| Heat shock protein (HSP) 90-beta | IPI00471584.5 | 1.50 ±0.09 | 9 | P34058 | 301252 |
| Heat-shock protein beta-1 | IPI00201586.1 | 2.55 ±0.24 | 4 | P42930 | 24471 |
| Heat-shock protein beta-8 | IPI00189624.1 | 2.09±NA | 1 | Q9EPX0 | 113906 |
| Hsc70-interacting protein | IPI00199273.1 | 1.83 ±0.05 | 3 | P50503 | 81800 |
| Hypoxia up-regulated 1 | IPI00559738.1 | 1.94±NA | 1 | Q63617 | 192235 |
| Kelch repeat and BTB domain-containing protein 10 | IPI00190417.1 | 9.96±NA | 1 | Q9ER30 | 117537 |
| Pincher | IPI00200271.1 | 3.01±NA | 1 | Q8R3Z7 | 192204 |
| PREDICTED: low density lipoprotein receptor-related protein associated protein 1 |
IPI00364124.1 | 1.56±NA | 1 | Q99068 | 116565 |
| PREDICTED: similar to CCT eta, eta subunit of the chaperonin containing TCP-1 |
IPI00364286.2 | 1.70 ±0.21 | 2 | 297406 | |
| T-complex protein 1 subunit alpha | IPI00200847.1 | 1.51±0.03 | 3 | P28480 | 24818 |
| Ubiquitin fusion degradation 1-like isoform 1 | IPI00195248.4 | 1.63±NA | 1 | Q9ES53 | 84478 |
| Cell-adhesion Proteins | |||||
| Glypican-1 precursor | IPI00194930.5 | 1.70 ±0.12 | 10 | P35053 | 58920 |
| Integrin beta-1 precursor | IPI00191681.1 | 2.30±NA | 1 | P49134 | 24511 |
| Lactadherin precursor | IPI00188896.1 | 2.32 ±0.17 | 8 | P70490 | 25277 |
| Neural cell adhesion molecule 1, 140 kDa isoform precursor |
IPI00476991.1 | 3.88±NA | 1 | P13596/Q3T1H3 | 24586 |
| PREDICTED: laminin, alpha 5 | IPI00190577.4 | 2.27±NA | 1 | P70636 | 140433 |
| PREDICTED: laminin, gamma 1 | IPI00363849.2 | 2.70±NA | 1 | P97552 | 117036 |
| PREDICTED: nidogen 2 | IPI00372786.3 | 2.43 ±0.33 | 9 | Q5M812 | 302248 |
| PREDICTED: similar to alpha 3 type VI collagen isoform 1 precursor |
IPI00360737.2 | 1.51 ±0.10 | 4 | 367313 | |
| PREDICTED: similar to Elastin microfibril interfacer 1 |
IPI00199867.1 | 2.83 ±0.25 | 7 | 298845 | |
| PREDICTED: similar to laminin B1 | IPI00365542.2 | 1.50 ±0.05 | 2 | 298941 | |
| PREDICTED: similar to Transmembrane 4 superfamily member 6 |
IPI00201753.1 | 3.18 ±0.00 | 2 | Q5RJZ3 | 302313 |
| PREDICTED: similar to type XV collagen | IPI00364868.2 | 3.21 ±0.45 | 4 | Q4G024 | 298069 |
| PREDICTED: similar to Vinculin | IPI00365286.3 | 1.55 ±0.12 | 17 | Q9ESQ3 | 305679 |
| Procollagen C-endopeptidase enhancer 1 precursor | IPI00194566.1 | 2.44 ±0.06 | 4 | O08628 | 29569 |
| Protein-lysine 6-oxidase precursor | IPI00214661.1 | 2.56±NA | 1 | P16636 | 24914 |
| Splice Isoform 1 of Fibronectin precursor | IPI00200757.1 | 2.30 ±0.17 | 50 | P04937 | 25661 |
| OX-2 membrane glycoprotein precursor(Cd200) | IPI00193967.2 | 5.26±NA | 1 | P04218 | 24560 |
| PREDICTED: similar to Fibulin-1 precursor | IPI00370411.2 | 1.92±NA | 1 | 315191 | |
| PREDICTED: similar to fibulin-2 | IPI00388257.3 | 1.55±0.90 | 8 | Q8CJG7 | 282583 |
| Cell Structure and Motility | |||||
| Actin, alpha cardiac | IPI00194087.3 | 1.61±0.13 | 10 | P68035 | 29275 |
| Actin, alpha skeletal muscle | IPI00189813.1 | 1.98 ±1.38 | 11 | P68136 | 29437 |
| Actin-related protein 2/3 complex subunit 1A | IPI00200845.1 | 2.19±NA | 1 | Q6PCU9 | 81824 |
| Microtubule-associated protein 1S | IPI00362631.1 | 1.81±NA | 1 | 290640 | |
| Double cortin and calcium/calmodulin-dependent protein kinase-like 1 |
IPI00373202.2 | 11.43±NA | 1 | O08875 | 83825 |
| General vesicular transport factor p115 | IPI00324618.3 | 1.57 ±0.20 | 2 | P41542 | 56042 |
| Hypothetical protein RGD1305887 | IPI00195673.1 | 2.09 ±1.41 | 15 | Q4QQV0 | 307351 |
| Isoform 1 of Tropomyosin alpha-3 chain(Tpm3) | IPI00372259.4 | 1.58±0.65 | 5 | Q63610 | 117557 |
| Kinesin-1 heavy chain | IPI00364904.2 | 4.43 ±0.52 | 10 | Q2PQA9 | 117550 |
| Microtubule-associated protein 1A | IPI00199693.2 | 1.76±NA | 1 | P34926 | 25152 |
| Microtubule-associated protein 4 | IPI00393975.2 | 1.71 ±0.09 | 4 | Q5M7W5 | 367171 |
| Microtubule-associated protein RP/EB family member 3 |
IPI00360288.1 | 3.35±NA | 1 | Q5XIT1 | 298848 |
| Myosin heavy chain, fast skeletal muscle, embryonic | IPI00201578.1 | 219.81 ±2.46 | 4 | P1284 | 24583 |
| Myosin light polypeptide 4 | IPI00214457.1 | 127.46±35.09 | 5 | P17209 | 688228 |
| Myosin, heavy polypeptide 2, skeletal muscle, adult | IPI00554308.2 | 443.05±NA | 1 | Q0GC40 | 691644 |
| Nestin | IPI00194103.1 | 7.23 ±0.84 | 8 | P21263 | 25491 |
| PREDICTED: similar to cofilin | IPI00369419.2 | 4.49±NA | 1 | P45592 | 366624 |
| PREDICTED: similar to cytoskeleton-associated protein 4 |
IPI00365982.1 | 1.66 ±0.17 | 6 | 362859 | |
| PREDICTED: similar to gamma-filamin | IPI00358175.2 | 4.90 ±0.28 | 8 | Q8VHX6(m) | 362332 |
| PREDICTED: similar to microfilament and actin filament cross-linker protein isoform a |
IPI00359003.3 | 3.07 ±0.48 | 2 | 362587 | |
| PREDICTED: similar to nebulin | IPI00372072.2 | 3.43±NA | 1 | 311029 | |
| PREDICTED: similar to titin isoform N2-B | IPI00564395.1 | 7.65±NA | 1 | P97850 | 84015 |
| Septin-7 | IPI00204899.1 | 1.52 ±0.14 | 4 | A2VCW8 | 64551 |
| Smooth muscle alpha-actin | IPI00197129.1 | 2.69±3.13 | 14 | Q63030 | 81633 |
| Spectrin alpha chain, brain | IPI00209258.4 | 1.57 ±0.09 | 18 | Q6IRK8 | 64159 |
| Splice Isoform 1 of Tropomyosin 1 alpha chain | IPI00197888.2 | 2.06 ±0.10 | 3 | P04692 | 24851 |
| Splice Isoform 2 of Tropomyosin beta chain | IPI00187731.4 | 2.75 ±0.55 | 3 | P58775 | 500450 |
| Stomatin (Epb7.2)-like 2 | IPI00203528.1 | 1.80±NA | 1 | Q4FZT0 | 298203 |
| Transgelin | IPI00231196.4 | 4.90 ±0.00 | 2 | O08564 | 25123 |
| Vesicle-associated membrane protein-associated protein A |
IPI00209290.2 | 2.25 ±0.02 | 5 | Q9Z270 | 58857 |
| Metabolism | |||||
| 2,4-dienoyl-CoA reductase, mitochondrial precursor | IPI00213659.3 | 1.94 ±0.19 | 3 | Q64591 | 117543 |
| 6-phosphofructokinase, muscle type | IPI00231293.6 | 1.57±NA | 1 | P47858 | 65152 |
| Aldose reductase | IPI00231737.4 | 1.65 ±0.04 | 3 | P07943 | 24192 |
| Alpha-enolase | IPI00464815.10 | 1.60 ±0.05 | 11 | P04764 | 24333 |
| Alpha-N-acetylglucosaminidase | IPI00370034.2 | 2.01±NA | 1 | Q5XIA5 | 287711 |
| Annexin A11 | IPI00364621.2 | 1.76±NA | 1 | Q5XI77 | 290527 |
| Asparagine synthetase | IPI00471908.5 | 4.10±NA | 1 | P49088 | 25612 |
| Aspartate aminotransferase, cytoplasmic | IPI00421513.6 | 1.63±NA | 1 | P13221 | 24401 |
| ATP synthase alpha chain, mitochondrial precursor | IPI00396910.1 | 1.93 ±0.29 | 5 | P15999 | 65262 |
| ATP synthase B chain, mitochondrial precursor | IPI00196107.1 | 2.23±0.00 | 2 | P19511 | 171375 |
| ATP synthase beta chain, mitochondrial precursor | IPI00551812.1 | 2.08 ±0.13 | 22 | P10719 | 171374 |
| ATP synthase delta chain, mitochondrial precursor | IPI00198620.1 | 2.04±0.05 | 2 | P35434 | 245965 |
| ATP synthase D chain, mitochondrial | IPI00230838.4 | 2.48 ±0.00 | 2 | P31399 | 641434 |
| ATP synthase, H+ transporting, mitochondrial F1 Complex, gamma polypeptide 1 |
IPI00396906.1 | 2.21±NA | 1 | Q6PCU0 | 116550 |
| Beta-hexosaminidase alpha chain precursor | IPI00394353.1 | 1.74±0.09 | 3 | Q641X3 | 300757 |
| Beta-hexosaminidase beta chain precursor | IPI00464518.1 | 1.55±NA | 1 | Q6AXR4 | 294673 |
| Biliverdin reductase A precursor | IPI00230874.10 | 7.20±NA | 1 | P46844 | 116599 |
| Citrate synthase | IPI00206977.1 | 2.10±0.33 | 2 | Q8VHF5 | 170587 |
| COX15 homolog, cytochrome c oxidase assembly protein |
IPI00361315.2 | 1.74±NA | 1 | Q3T1G9 | 309391 |
| Cytochrome c oxidase polypeptide Va, mitochondrial precursor |
IPI00192246.1 | 2.48 ±0.03 | 3 | P11240 | 252934 |
| Cytochrome c, somatic | IPI00231864.4 | 1.90±0.03 | 3 | P62898 | 25309 |
| D-3-phosphoglycerate dehydrogenase | IPI00475835.2 | 1.60 ±0.00 | 2 | O08651 | 58835 |
| Dihydrolipoamide dehydrogenase | IPI00365545.1 | 2.64 ±0.16 | 2 | Q6P6R2 | 298942 |
| Dihydrolipoamide S-acetyltransferase | IPI00231714.3 | 1.67 ±0.66 | 4 | P08461 | 81654 |
| Electron transfer flavoprotein beta-subunit | IPI00364321.2 | 1.91±NA | 1 | Q68FU3 | 292845 |
| Fructose-bisphosphate aldolase A | IPI00231734.4 | 3.16 ±0.10 | 13 | P05065 | 24189 |
| Fumarate hydratase, mitochondrial precursor | IPI00231611.7 | 1.91 ±0.85 | 4 | P14408 | 24368 |
| Glucose phosphate isomerase | IPI00364311.1 | 2.09±0.10 | 4 | Q6P6V0 | 292804 |
| Glucosidase, alpha acid | IPI00400579.1 | 1.61 ±0.65 | 5 | Q6P7A9 | 367562 |
| Glyceraldehyde-3-phosphate dehydrogenase | IPI00212647.2 | 2.76 ±0.19 | 10 | P04797 | 24383 |
| GTP:AMP phosphotransferase mitochondrial | IPI00362243.6 | 2.38 ±0.06 | 2 | P29411 | 26956 |
| Hsd17b4 protein | IPI00326948.2 | 1.66 ±0.00 | 2 | Q6IN39 | 79244 |
| Hypothetical LOC361596 | IPI00464897.1 | 1.53 ±0.17 | 4 | Q6DGF1 | 361596 |
| Hypothetical protein LOC360975 | IPI00215093.1 | 1.66 ±0.08 | 4 | Q5XI78 | 360975 |
| Isocitrate dehydrogenase [NAD] subunit beta, mitochondrial precursor |
IPI00357924.1 | 1.51 ±0.25 | 4 | Q68FX0 | 94173 |
| Lactoylglutathione lyase | IPI00188304.2 | 1.53±0.01 | 3 | Q6P7Q4 | 294320 |
| L-lactate dehydrogenase A chain | IPI00197711.1 | 2.10±0.11 | 6 | P04642 | 24533 |
| Low molecular mass ubiquinone-binding protein | IPI00382312.3 | 2.36±NA | 1 | Q7TQ16 | 497902 |
| LRRGT00113 | IPI00196629.3 | 2.00±NA | 1 | 499358 | |
| Malate dehydrogenase, mitochondrial | IPI00566583.1 | 2.04±0.09 | 8 | 81829 | |
| Methylmalonate-semialdehyde dehydrogenase [acylating], mitochondrial precursor |
IPI00205018.2 | 4.11±NA | 1 | Q02253 | 81708 |
| Mitochondrial-processing peptidase alpha subunit, mitochondrial |
IPI00195551.1 | 2.06 ±0.05 | 3 | P20069 | 296588 |
| N-acylglucosamine 2-epimerase | IPI00204162.1 | 2.17±0.00 | 2 | P51607 | 81759 |
| NADH-cytochrome b5 reductase | IPI00231662.5 | 1.50 ±0.07 | 8 | P20070 | 25035 |
| Nicotinamide nucleotide transhydrogenase | IPI00555265.1 | 1.90±0.26 | 2 | Q5BJZ3 | 310378 |
| Nucleoside diphosphate kinase A | IPI00194404.5 | 1.54 ±0.05 | 7 | Q05982 | 191575 |
| Peptidase D | IPI00364304.2 | 1.78±NA | 1 | Q5I0D7 | 292808 |
| PREDICTED: dihydrolipoamide branched chain transacylase E2 |
IPI00373418.3 | 1.96±0.00 | 2 | Q99PU6 | 29611 |
| PREDICTED: similar to catechol-O-methyltransferase domain containing 1 |
IPI00365293.2 | 1.98±NA | 1 | 305685 | |
| PREDICTED: similar to Cox7a2l protein | IPI00365505.2 | 2.49±NA | 1 | P28075 | 316064 |
| PREDICTED: similar to Cytochrome c oxidase polypeptide VIb |
IPI00389152.3 | 1.68 ±0.11 | 4 | 502592 | |
| PREDICTED: similar to glyceraldehyde-3-phosphate dehydrogenase |
IPI00554039.1 | 2.78 ±0.21 | 10 | 498099 | |
| PREDICTED: similar to Phosphoacetylglucosamine mutase |
IPI00205603.3 | 2.80 ±0.06 | 2 | 363109 | |
| PREDICTED: similar to pyrroline-5-carboxylate synthetase short isoform |
IPI00372524.3 | 1.52 ±0.32 | 9 | 361755 | |
| PRx III (Thioredoxin-dependent peroxide reductase, mitochondrial) |
IPI00208215.1 | 2.01 ±0.07 | 3 | Q9Z0V6 | 64371 |
| Pyruvate dehydrogenase E1 component alpha subunit, somatic form, mitochondrial precursor |
IPI00191707.3 | 4.27 ±4.91 | 3 | P26284 | 29554 |
| Pyruvate dehydrogenase E1 component subunit beta, mitochondrial precursor |
IPI00194324.1 | 2.91 ±1.75 | 3 | P49432 | 289950 |
| Serum deprivation response protein | IPI00362416.1 | 3.41±NA | 1 | Q66H98 | 316384 |
| Superoxide dismutase [Mn], mitochondrial precursor | IPI00211593.1 | 1.57 ±0.00 | 2 | P07895 | 24787 |
| Trifunctional enzyme alpha subunit, mitochondrial precursor |
IPI00212622.1 | 1.71 ±0.08 | 10 | Q64428 | 170670 |
| Trifunctional enzyme beta subunit, mitochondrial precursor |
IPI00198467.1 | 1.66±NA | 1 | Q60587 | 171155 |
| Triosephosphate isomerase | IPI00231767.4 | 3.72 ±0.91 | 2 | P48500 | 24849 |
| Ubiquinol-cytochrome c reductase core protein I | IPI00471577.1 | 2.52 ±0.36 | 4 | Q68FY0 | 301011 |
| Ubiquinol-cytochrome c reductase core protein II | IPI00480805.1 | 2.78±0.54 | 4 | P32551 | 293448 |
| PREDICTED: similar to cytochrome c-1 | IPI00366416.1 | 2.60 ±0.56 | 4 | 300047 | |
| SIALIDASE-1 PRECURSOR. | IPI00201456.5 | 2.07±NA | 1 | Q99PW3 | 24591 |
| Fumarylacetoacetate hydrolase domain-containing protein 1 |
IPI00368708.2 | 2.54±NA | 1 | Q6AYQ8 | 302980 |
| Long-chain-fatty-acid--CoA ligase 1 | IPI00188989.1 | 4.29 ±1.17 | 3 | P18163 | 25288 |
| Phosphoglycerate kinase 1 | IPI00231426.5 | 2.17 ±0.16 | 7 | P16617 | 24644 |
| PREDICTED: similar to inosine monophosphate dehydrogenase 1 isoform b |
IPI00480747.2 | 1.72±NA | 1 | 362329 | |
| PREDICTED: similar to NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 1 |
IPI00373108.2 | 2.44±NA | 1 | 296086 | |
| Pyruvate dehydrogenase [lipoamide] kinase isozyme 1, mitochondrial precursor |
IPI00204957.1 | 4.57±NA | 1 | Q63065 | 116551 |
| Sulfated glycoprotein 1 precursor | IPI00195160.1 | 1.72 ±0.32 | 2 | P10960 | 25524 |
| Acyl-Coenzyme A thioesterase 2, mitochondrial | IPI00358498.2 | 1.64 ±0.00 | 2 | O55171 | 302640 |
| PREDICTED: similar to Myotubularin-related protein 2 |
IPI00362271.3 | 1.61±NA | 1 | 315422 | |
| PREDICTED: similar to phosphoenolpyruvate carboxykinase 2 |
IPI00388232.3 | 1.77 ±0.43 | 2 | 361042 | |
| Serine hydroxymethyl transferase 2 | IPI00195109.3 | 1.80 ±0.08 | 2 | Q5U3Z7 | 299857 |
| Splice Isoform 1 of Lipid phosphate phosphohydrolase 1 |
IPI00193763.1 | 4.59±NA | 1 | O08564 | 64369 |
| Long-chain-fatty-acid--CoA ligase 3 | IPI00205908.1 | 3.25 ±0.70 | 3 | Q63151 | 114024 |
| NAD(P)H:quinone oxidoreductase type 3, polypeptide A2 |
IPI00371971.2 | 2.29±0.28 | 2 | Q5EB81 | 304805 |
| PREDICTED: similar to Pyruvate dehydrogenase kinase, isoenzyme 3 |
IPI00192133.3 | 1.98±NA | 1 | 296849 | |
| Adenylate kinase isoenzyme 4, mitochondrial | IPI00204311.1 | 6.46±NA | 1 | Q9WUS0 | 29223 |
| NADH dehydrogenase subunit 4 | IPI00200487.1 | 1.55±NA | 1 | P05508 | |
| acyl-CoA thioesterase 7 | IPI00213571.1 | 1.84±NA | 1 | Q64559 | 26759 |
| Dehydrogenase/reductase (SDR family) member 7B | IPI00369545.2 | 3.09±NA | 1 | Q5RJY4 | 287380 |
| 11 kDa protein | IPI00390086.2 | 1.83±NA | 1 | Q6IBB3(h) | 690441 |
| Ubiquinol-cytochrome c reductase complex 11 kDa protein, mitochondrial precursor |
IPI00369093.1 | 2.48±NA | 1 | Q5M9I5 | 366448 |
| 20alpha-hydroxysteroid dehydrogenase | IPI00189189.2 | 23.55±NA | 1 | Q91XV8 | 171516 |
| NADH dehydrogenase (Ubiquinone) Fe-S protein 1 | IPI00358033.1 | 1.67±NA | 1 | Q66HF1 | 301458 |
| 87 kDa protein | IPI00212665.2 | 1.76 ±0.12 | 2 | 116645 | |
| Similar to CG6105-PA | IPI00421711.1 | 2.58±NA | 1 | Q6PDU7 | 300677 |
| Hypothetical protein LOC314432 | IPI00368347.2 | 1.63 ±0.04 | 8 | Q5U300 | 314432 |
| Transporter or Channel | |||||
| ADP/ATP translocase 2 | IPI00200466.2 | 1.78 ±0.28 | 3 | Q09073 | 25176 |
| ATPase, H+ transporting, V1 subunit E isoform 1 | IPI00400615.1 | 1.63 ±1.16 | 2 | Q6PCU2 | 297566 |
| Cationic amino acid transporter-1 | IPI00190498.1 | 1.81±NA | 1 | Q8VIA9 | 25648 |
| Fragile X mental retardation gene 1, autosomal homolog |
IPI00373184.2 | 3.19 ±0.34 | 3 | Q5XI81 | 361927 |
| Mitochondrial 2-oxoglutarate/malate carrier protein | IPI00231261.6 | 1.97±NA | 1 | P97700 | 64201 |
| Na-K-Cl cotransporter | IPI00212590.1 | 2.34±NA | 1 | Q9QX10 | 83629 |
| Nuclear protein localization protein 4 homolog | IPI00191492.2 | 1.51±NA | 1 | Q9ES54 | 140639 |
| PREDICTED: secretory carrier membrane protein 3 | IPI00206037.5 | 1.52±NA | 1 | Q9ERM8 | 65169 |
| PREDICTED: similar to ATPase, H+ transporting, V1 subunit A, isoform 1 |
IPI00373076.1 | 1.86 ±0.41 | 2 | 360716 | |
| PREDICTED: similar to importin 7 | IPI00206234.2 | 1.89 ±0.62 | 3 | 308939 | |
| PREDICTED: similar to p59 immunophilin | IPI00358443.2 | 1.59±NA | 1 | Q8K3U8 | 260321 |
| PREDICTED: similar to Tweety homolog 2 | IPI00361325.2 | 3.49±NA | 1 | 287803 | |
| Rho/rac guanine nucleotide exchange factor | IPI00368617.2 | 1.66 ±0.05 | 3 | Q5FVC2 | 310635 |
| Sideroflexin-1 | IPI00213735.2 | 9.59±NA | 1 | Q63965 | 364678 |
| Similar to SEC24 related gene family, member | IPI00365299.2 | 1.55 ±0.21 | 3 | 685144 | |
| Slc25a3 protein | IPI00209115.2 | 2.51 ±0.03 | 5 | Q6IRH6 | 245959 |
| Sodium- and chloride-dependent taurine transporter | IPI00327953.2 | 2.67±0.07 | 2 | P31643 | 29464 |
| Splice Isoform SERCA2B of Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 |
IPI00190020.3 | 2.20 ±0.00 | 2 | Q71UZ2 | 29693 |
| Splice Splice Isoform IIBA of Dynamin-2 | IPI00210319.2 | 1.59±NA | 1 | P39052 | 25751 |
| Vacuolar ATP synthase subunit B, brain isoform | IPI00199305.1 | 1.96 ±0.11 | 5 | P62815 | 117596 |
| Vacuolar ATP synthase subunit F | IPI00198291.1 | 1.67±NA | 1 | P50408 | 116664 |
| PREDICTED: similar to peroxisomal integral membrane protein |
IPI00366455.1 | 1.86±NA | 1 | 300083 | |
| Cell Cycle,Cell Proliferation and Differentiation | |||||
| Casein kinase II subunit alpha | IPI00192586.1 | 1.69±NA | 1 | P19139 | 116549 |
| NG,NG-dimethylarginine dimethylaminohydrolase 1 | IPI00231194.4 | 1.81±NA | 1 | O08557 | 64157 |
| Nucleosome assembly protein 1-like 4 | IPI00366110.3 | 1.53 ±0.05 | 2 | 361684 | |
| PREDICTED: similar to cullin 4A | IPI00364684.2 | 1.61±NA | 1 | 361181 | |
| PREDICTED: similar to hepatic multiple inositol polyphosphate phosphatase |
IPI00364031.1 | 2.69 ±0.00 | 2 | 499084 | |
| Single-stranded DNA-binding protein, mitochondrial precursor |
IPI00196750.1 | 1.82 ±0.20 | 2 | P28042 | 54304 |
| Others and Molecular Function Unclassified | |||||
| 37 kDa protein | IPI00562745.1 | 2.94±NA | 1 | ||
| 9 kDa protein | IPI00567137.1 | 2.28 ±0.77 | 2 | ||
| Coiled-coil domain-containing protein 47 precursor | IPI00203647.1 | 1.92 ±0.00 | 2 | Q5U2X6 | 303606 |
| Cold shock domain-containing protein E1 | IPI00190971.1 | 1.62±0.00 | 2 | P18395 | 117180 |
| Complement component 1, Q subcomponent-binding protein, mitochondrial precursor |
IPI00361686.4 | 2.26 ±0.15 | 4 | O35796 | 29681 |
| Cysteine-rich with EGF-like domain protein 1 precursor |
IPI00202520.1 | 2.87±NA | 1 | Q4V7F2 | 312638 |
| FK506-binding protein 9 precursor | IPI00215190.1 | 1.57 ±0.00 | 2 | Q66H94 | 297123 |
| Hypothetical protein | IPI00389960.2 | 1.87±NA | 1 | Q5XI01 | 686883 |
| Hypothetical protein LOC498174 | IPI00394488.2 | 1.84±NA | 1 | Q5RK08 | 498174 |
| Hypothetical protein RGD1306649 | IPI00197896.1 | 2.46 ±0.20 | 2 | Q4FZT8 | 288772 |
| Kidney predominant protein NCU-G1 | IPI00196226.1 | 1.55 ±0.00 | 2 | Q68FV6 | 295231 |
| LOC500199 protein | IPI00204675.4 | 1.60±NA | 1 | Q4KM70 | 500199 |
| Major vault protein | IPI00231381.7 | 1.92 ±0.03 | 2 | Q62667 | 64681 |
| Myeloid-associated differentiation marker | IPI00339007.1 | 2.17±NA | 1 | Q6VBQ5 | 369016 |
| N-acylsphingosine amidohydrolase 1 | IPI00421601.3 | 1.81 ±0.02 | 2 | Q9EQJ6 | 84431 |
| Nicalin precursor | IPI00369465.2 | 2.15±NA | 1 | Q5XIA1 | 314648 |
| Nucleolar protein 3 | IPI00209297.1 | 2.35 ±0.08 | 2 | Q62881 | 85383 |
| O-GlcNAcase | IPI00208152.1 | 17.41±NA | 1 | Q8VIJ5 | 154968 |
| Paraspeckle protein 1 | IPI00203753.1 | 1.52±NA | 1 | Q4KLH4 | 305910 |
| Peroxisomal biogenesis factor 11b | IPI00210003.1 | 2.31±NA | 1 | Q4KM24 | 310682 |
| Pleckstrin homology domain containing, family C | IPI00362106.2 | 1.99 ±0.14 | 3 | Q5XI19 | 289992 |
| Poliovirus receptor | IPI00326594.11 | 8.10±NA | 1 | 25066 | |
| PREDICTED: ATPase, H+ transporting, lysosomal accessory protein 2 |
IPI00358308.2 | 2.24±NA | 1 | Q6AXS4 | 302526 |
| PREDICTED: similar to 0910001A06Rik protein | IPI00366405.2 | 1.87 ±0.00 | 2 | 299909 | |
| PREDICTED: similar to ARL6IP2 | IPI00365499.3 | 2.17±NA | 1 | Q562A0 | 298757 |
| PREDICTED: similar to C21ORF80 | IPI00360543.2 | 1.79 ±0.00 | 2 | 309686 | |
| PREDICTED: similar to Coiled-coil-helix-coiled-coil-helix domain containing 6 |
IPI00364520.2 | 2.04 ±0.00 | 2 | 297436 | |
| PREDICTED: similar to collagen alpha 1(IV) chain precursor - mouse |
IPI00362887.2 | 1.57 ±0.00 | 3 | Q5FWY9 | 290905 |
| PREDICTED: similar to GPI transamidase component PIG-T precursor |
IPI00373349.1 | 1.50 ±0.00 | 2 | 296360 | |
| PREDICTED: similar to heparan sulfate proteoglycan 2 |
IPI00388323.4 | 2.33 ±0.21 | 24 | Q62980 | 117511 |
| PREDICTED: similar to Histidine triad nucleotide-binding protein 2 |
IPI00358757.2 | 1.81±0.00 | 2 | 313491 | |
| PREDICTED: similar to mKIAA0312 protein | IPI00196914.4 | 1.58±0.04 | 3 | P51593 | 501546 |
| PREDICTED: similar to NEDD9 interacting protein with calponin homology and LIM domains |
IPI00371967.2 | 2.10±NA | 1 | 294520 | |
| PREDICTED: similar to Pre-B-cell leukemia transcription factor interacting protein 1 |
IPI00201858.3 | 4.00 ±4.69 | 2 | A2VD12 | 310644 |
| PREDICTED: similar to RIKEN cDNA 0710008K08 |
IPI00210521.2 | 2.27±0.00 | 2 | 361185 | |
| PREDICTED: similar to RIKEN cDNA 5033414D02 |
IPI00209463.2 | 1.90±NA | 1 | 293888 | |
| PREDICTED: similar to RIKEN cDNA 5730434I03 gene |
IPI00364212.2 | 1.77 ±0.00 | 2 | 305284 | |
| PREDICTED: similar to signal recognition particle,72 kDa subunit |
IPI00565085.1 | 1.64±0.14 | 3 | 499086 | |
| PREDICTED: similar to Transcriptional activator protein PUR-alpha |
IPI00197411.1 | 1.62 ±0.09 | 4 | 307498 | |
| Procollagen-lysine,2-oxoglutarate 5-dioxygenase 3 precursor |
IPI00331772.4 | 1.97 ±0.22 | 3 | Q5U367 | 288583 |
| Proliferation related acidic leucine rich protein PAL31 |
IPI00192336.1 | 1.68±NA | 1 | Q9EST6 | 170724 |
| Protein FAM98A | IPI00554081.1 | 1.73±NA | 1 | Q5FWT1 | 313873 |
| Protein KIAA0152 precursor | IPI00371173.2 | 2.32 ±0.00 | 2 | Q5FVQ4 | 304543 |
| PTPRF interacting protein, binding protein 1 (liprin beta 1) |
IPI00567984.1 | 1.63±0.00 | 2 | 312855 | |
| Reticulon 3, isoform A1 | IPI00421506.1 | 1.64 ±0.20 | 4 | A1L1I6 | 140945 |
| Secreted acidic cysteine rich glycoprotein | IPI00557175.1 | 2.54±0.15 | 7 | P16975 | 24791 |
| Similar to PRUNEM1 | IPI00368646.1 | 3.17±NA | 1 | Q6AYG3 | 310664 |
| Splice Isoform 2 of Procollagen-lysine,2-oxoglutarate 5-dioxygenase 2 precursor |
IPI00208888.1 | 1.74 ±0.32 | 4 | Q811A3 | 300901 |
| Tangerin | IPI00203791.2 | 4.55±NA | 1 | Q5PQM3 | 309169 |
| Thymosin beta-10 | IPI00231695.5 | 2.61±NA | 1 | P63312 | 50665 |
| Transmembrane emp24 protein transport domain containing 9 |
IPI00364707.1 | 1.91 ±0.00 | 2 | Q5I0E7 | 361207 |
| Transmembrane protein 109 precursor | IPI00372499.1 | 2.21±NA | 1 | Q6AYQ4 | 361732 |
| LOC360721 protein | IPI00608138.1 | 2.87±NA | 1 | Q4QQV3 | 360721 |
| Up-regulated during skeletal muscle growth protein 5 |
IPI00202111.1 | 2.28 ±0.00 | 2 | Q9JJW3 | 171069 |
| WD-containing protein | IPI00205633.6 | 2.30±NA | 1 | Q9R037 | 246152 |
Table 2.
The List of Proteins with the Ratio ≤05
| Description | IPI ID | Average Ratio Average±SD |
Quantification Peptide Num |
UniProtKB ID/AC |
Gene ID |
|---|---|---|---|---|---|
| Alcohol dehydrogenase class 4 mu/sigma chain | IPI00324743.1 | 0.38 ±0.03 | 3 | P41682 | 171178 |
| Amidophosphoribosyltransferase precursor | IPI00198619.1 | 0.43±NA | 1 | P35433 | 117544 |
| APEX (Fragment) | IPI00200918.1 | 0.48 ±NA | 1 | Q99PF3 | 79116 |
| Capping protein | IPI00464670.1 | 0.39 ±0.00 | 2 | Q6AYC4 | 297339 |
| CD14 antigen | IPI00231949.5 | 0.24±NA | 1 | Q63691 | 60350 |
| Galectin-3 | IPI00194341.4 | 0.44 ±0.01 | 5 | P08699 | 83781 |
| Glutaredoxin-1 | IPI00231191.6 | 0.49 ±0.07 | 2 | Q9ESH6 | 64045 |
| Mevalonate kinase | IPI00214563.1 | 0.20 ±NA | 1 | P17256 | 81727 |
| Minichromosome maintenance protein 7 | IPI00371012.2 | 0.49±0.07 | 4 | Q1PS21 | 288532 |
| Mtap6 protein (Fragment) | IPI00212447.2 | 0.31±NA | 1 | Q6AYX8 | 29457 |
| N-myc downstream regulated gene 1 | IPI00421389.1 | 0.38±NA | 1 | Q6JE36 | 299923 |
| NORBIN | IPI00205396.1 | 0.44±NA | 1 | O35095 | 89791 |
| Pleckstrin homology-like domain, family B, member 1 |
IPI00207827.2 | 0.36±NA | 1 | Q63312 | 171434 |
| PREDICTED: cadherin 11 | IPI00211883.2 | 0.43 ±0.02 | 2 | Q9JIW2 | 84407 |
| PREDICTED: similar to 2610304F09Rik protein | IPI00358454.2 | 0.46 ±0.00 | 2 | 307527 | |
| PREDICTED: similar to Coronin, actin binding protein 1C |
IPI00388015.3 | 0.33±0.46 | 3 | B2RYG0 | 501841 |
| PREDICTED: similar to DNA replication licensing factor MCM3 |
IPI00361669.2 | 0.44±NA | 1 | 367976 | |
| PREDICTED: similar to DNA replication licensing factor MCM3 |
IPI00371781.2 | 0.44±NA | 1 | 316273 | |
| PREDICTED: similar to Filamin B | IPI00373752.2 | 0.47 ±0.12 | 6 | 306204 | |
| PREDICTED: similar to Lmnb2 protein | IPI00366190.3 | 0.42 ±NA | 1 | 299625 | |
| PREDICTED: similar to Stromal cell-derived factor 2 precursor |
IPI00369655.1 | 0.38±NA | 1 | 287470 | |
| PREDICTED: similar to TF-1 apoptosis related protein 19 |
IPI00193547.2 | 0.33 ±NA | 1 | 292814 | |
| PREDICTED: similar to Tripartite motif protein 47 | IPI00361534.1 | 0.3±NA | 1 | 690374 | |
| PREDICTED: similar to Williams-Beuren syndrome deletion transcript 9 homolog |
IPI00370333.1 | 0.29±NA | 1 | Q2V6G6 | 368002 |
| Prostaglandin F2 receptor negative regulator precursor |
IPI00208280.2 | 0.47 ±0.27 | 3 | Q62786 | 29602 |
| prostaglandin-endoperoxide synthase 1 | IPI00567836.1 | 0.48±0.01 | 3 | Q66HK3 | 24693 |
| Proteasome inhibitor PI31 subunit | IPI00391791.2 | 0.27±NA | 1 | Q5XIU5 | 682071 |
| Reck reversion-inducing-cysteine-rich protein with kazal motifs |
IPI00358750.2 | 0.48 ±NA | 1 | 313488 | |
| Ribonucleotide reductase M1 | IPI00361151.2 | 0.47±NA | 1 | Q5U2Q5 | 365320 |
| RNA terminal phosphate cyclase domain 1 | IPI00201443.1 | 0.37 ±NA | 1 | Q68FS8 | 295395 |
| RNA-binding motif protein 3 | IPI00367437.3 | 0.50 ±0.00 | 2 | Q925G0 | 114488 |
| Small nuclear ribonucleoprotein polypeptide A | IPI00364044.3 | 0.20 ±0.00 | 2 | Q5U214 | 292729 |
| Testis derived transcript | IPI00560148.2 | 0.4±NA | 1 | Q2LAP6 | 500040 |
| Transcriptional repressor CTCF | IPI00207172.2 | 0.47±NA | 1 | Q9R1D1 | 83726 |
| UDP-glucose 6-dehydrogenase | IPI00195803.1 | 0.44±0 | 2 | O70199 | 83472 |
| Wiskott-Aldrich syndrome protein-interacting protein |
IPI00470232.5 | 0.3±NA | 1 | Q6IN36 | 117538 |
| Xanthine dehydrogenase/oxidase | IPI00231694.6 | 0.50 ±0.07 | 3 | P22985 | 497811 |
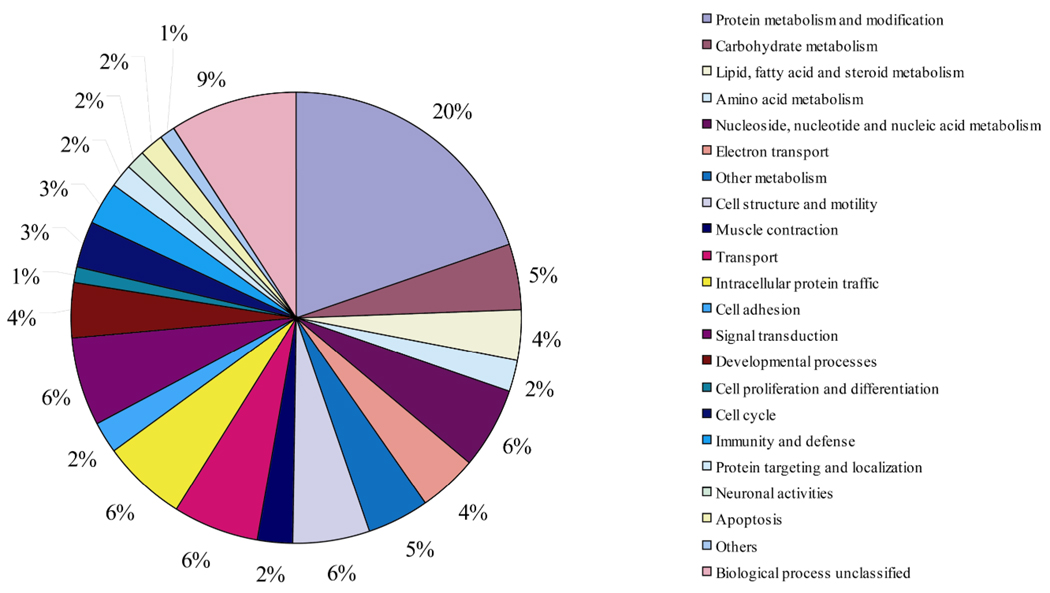
Figure 3.
Functional annotations of the proteins with > 1.5 ratio in L6 myotubes compared to myoblasts. The pie chart shows the distribution of biological processes to all up regulated proteins in L6 myotubes compared to myoblasts in this study using panther classification system with modification.
3.3. The comparative profiling of protein expression
Many of the differentially expressed proteins observed in this study have been reported to be involved in myogenic regulation, and some were newly discovered by this study.
Proteins increased in expression (ratio ≥1.5) were sorted by the PANTHER Classification system (Figure 3 and Table 1). These proteins are mainly involved in inter-or intracellular signaling, protein synthesis and protein degradation, protein folding, cell adhesion and extracellular matrix, cell structure and motility, metabolism, substance transportation, etc. These patterns of up-regulation were consistent with the functional and structural characteristics of skeletal muscle cells. In order to explore functional modules of the proteins we quantified, protein interaction network was predicted by STRING (see supplementary Figure 2).
Mediators of signaling pathway
Myogenic differentiation is regulated by positive and negative signals from surroundings. After switching the cells from nutrient rich media to nutrient poor medium by lowering FBS content from 10% to 2%, L6 myoblasts are able to sense the physical and chemical signals of lowered FBS through specific membrane receptors [2]. Once L6 myoblasts sense these signals, a series of intracellular events will be triggered. The final result of these events is increased expression of MyoD and Myogenin. MyoD and Myogenin initiate multiple muscle differentiation-specific genes transcription for myogenic process [1, 2]. Although the role of mitogen-activated protein kinase (MAPK) signaling cascades in myogenesis is controversial, accumulating studies have shown that MAPK is activated during the differentiation of myogenic cell lines and is essential for the expression of muscle-specific genes [15, 16]. Activation of MAPK signaling cascades in myoblasts can modulate the activity of MyoD establishing dynamic modulation of the MyoD-induced programs of gene expression [3]. We observed that terminally differentiated myotubes increase the expression of some MAPK-pathway associated proteins, for example, Map4k4, MAPKK 1, etc. But, the amount of most MAPKs remains stable, because they are modified as functional executors at different stages of differentiation [17]. A kinase (PRKA) anchor protein 2, exhibiting protein kinase A binding, involved in actin filament organization, protein localization and the trans-membrane receptor protein serine/threonine kinase signaling pathway. Interestingly, prohibitin (PHB), a ubiquitously expressed and evolutionarily highly conserved protein, was found up-regulated once myoblasts initiated differentiation. This result is supported by western blot data (figure 2b) and the results from Tannu [6]. PHB has been found to be presented in the nucleus, the mitochondria and the plasma membrane. Gamble et al. reported that PHB participates in the activation of the Raf-MEK-ERK pathway [18, 19]. Sun and colleagues reported that PHB-2 can repress muscle differentiation by inhibiting MyoD and MEF2 in C2C12 cells [20]. From these clues, prohibitin may belong to MAPKs cascade and play important role in muscle differentiation. Besides MAPK related factors, we also found some other signaling molecules were up-regulated in muscle cells, such as inositol 1, 4, 5-triphosphate receptor 3 (IP3R-3), latent transforming growth factor beta binding protein 1 (LTBP-1), phosphatidylethanolamine-binding protein, SH2-containing inositol phosphatase 2 (SHIP2), and guanine nucleotide-binding protein G (i), alpha-2, etc. IP3R-3, in connection with acetylcholine signaling, adrenergic signaling, endothelin signaling, PDGF signaling, chemokine and cytokine signaling and Wnt signaling etc, is the receptor for inositol 1,4,5-trisphosphate to mediates the release of intracellular calcium. The alteration of IP3R-3 abundance in muscles may be so as to match the excitation-contraction coupling of muscle cell. LTBP-1 targets latent complexes of transforming growth factor beta to the extracellular matrix. It interacts with architectural extracellular matrix macromolecules—fibrillins that form ubiquitous extracellular microfibril suprastructures in the connective tissue space. It is unknown whether or not LTBP-1 participates in myogenic initiation and myoblast fusion [21, 22]. In addition, we identified one non-muscle differentiation-promoting protein, transcriptional activator protein Pur-beta (3.76 fold). This protein is known to regulate myeloid cell differentiation. It remains unclear how these proteins function in myogenic differentiation. In our study, many myogenesis-control factors, such as MyoD and Myogenin, were not observed, probably because these proteins were very low abundance in cells.
Protein Synthesis-and Degradation-related Proteins
Synthesis of Muscle-specific protein increases significantly during the myogensis process. Many proteins associated with proteins synthesis were found up-regulated in this study, including many amino acid-tRNA synthetases, eukaryotic translation elongation factors, eukaryotic translation initiation factors and ribosomal proteins. These types of proteins also have been reported by previous studies [6–9]. Protein synthesis and degradation is a finely coordinate process [23]. It is well known that the protein degradation system serves as a quality-control system for abnormal proteins to maintain cellular homeostasis [24]. And yet, as early as 1997, Gardrat hypothesized that ubiquitin-proteasome pathway was involved in muscle cell differentiation [25]. In 2005, Schwartz group discovered that both MyoD and inhibitor of DNA binding 1(Id1) are rapidly degraded by the ubiquitin-proteasome pathway during the differentiation of myoblast to myotube in mouse C2C12 myoblast cells, but the reduction of Id1 is more than MyoD markedly [26]. Rapid reduction of Id1 can release repression on MyoD, then, which will trigger muscle-specific gene transcription. This shows that ubiquitin-proteasome pathway is essential to initiation of mygenic differentiation by controlling muscle differentiation-specific gene expression [26]. Proteasomes, performing ATP-dependent proteolysis, are large protein complexes formed by many subunits. In this study, we found many proteasomal proteins, such as PSMA1, PSMA 2, PSMA 3, PSMA 4, PSMA5, PSMB1, PSMB2, PSMD2, PSMD3, the 26S protease regulatory subunit 7 and 26S protease regulatory subunit 4, were up-regulated during myogenic process. From the STRING network view, it can be seen directly that these proteins have the strong interactions (supplemented figure 2). Obviously, the changes of proteolytic system we found support the theory of myogenesis addressed by preceding publications [25, 26].
Molecular Chaperone
Molecular chaperones are a group of proteins whose roles are to assist newly translated proteins to fold properly as functional mature proteins or lead the misfolded proteins to degradation mentioned above. In differentiating muscle cells, the single nascent myosin molecule must go through folding and assemble into motor thick filament with associated proteins. It has been reported that Hsp90 and Hsc70 forms a complex with newly synthesized myosin and these chaperones promote myofibril assembly [27]. In this study, Hsp90, Hsc70 (heat shock cognate 71 kDa protein), Hsc70-interacting protein, have been up-regulated by 1.50, 1.58 and 1.83 fold respectively in L6 myotube cells. In addition, glucose-regulated protein precursor, hypoxia up-regulated 1 and heat shock 70 kDa protein 4, belonging to Hsp70 family chaperone, were also up-regulated in our study. That Hsp70 were increased during differentiation of myotubes has been proved by western blot [28]. T-complex protein 1 subunit alpha (TCP-1-alpha, CCT-alpha), a molecular chaperone of actin and tubulin, has also been found to be up-regulated by 1.7 fold. CCT activity is required for cell cycle progression and cytoskeleton organization in mammalian cells [29]. In this study, we also identified some small heat shock proteins, such as HspB1 (Hsp27), HspB8 (Hsp22), and aB-crystallin. These proteins can confer resistance to apoptosis during myogenic differentiation [30]. In addition, Hsp27 controlled by P38/MAPK pathway can modify actin polymerization. These behaviors of such proteins are beneficial to myogenic differentiation. Hsp27 and Hsp22 were up-regulated 2.55 and 2.09 fold respectively. Alpha B-crystallin also has important effect on myotubes development. This protein was identified in our experiment with no quantified SILAC ratio here. But, with manually check the MS spectra intensities and integral area of peptides of αB-crystallin, we found that αB-crystallin was up-regulated during myogenic process (Data not shown). In sum, the observed up-regulated molecular chaperones of cytoskeleton proteins play the important role in the muscle differentiation.
Cell-adhesion Proteins
Myoblasts-myotubes conversion requires cell-cell mutual interaction and fusion between myoblalsts. No doubt, adhesion molecules must be involved in this process. Some cell adhesion molecules have been reported to be involved in controlling the fusion of myoblasts during muscle development [31, 32]. Grossi and colleague have shown that mechanical stimulation can promote C2C12 cells differentiation through the laminin receptor [33]. In this study many extracellular matrix linker proteins were identified and showed increased expression in the L6 terminal differentiation stage, such as integrin beta-1 precursor (2.3 fold), isoform 1 of fibronectin precursor (2.3 fold), splice isoform 2 of fibronectin precursor (2.3 fold), procollagen C-proteinase enhancer protein (2.44 fold), protein-lysine 6-oxidase precursor (2.56 fold), splice isoform short of collagen alpha-1(XII) chain (1.73 fold) and vinculin (1.55 fold).. These proteins are involved in cell adhesion, cell communication, cell motility, and maintenance of cell shape. Integrin beta-1 is a subunit of several integrin proteins. Integrin is a receptor for fibronectin, collagen, and laminins. Brzoska has shown that integrin α3 subunit participates in myoblast adhesion and fusion in vitro [34]. When α3β1 integrin binds to its ligands, intracellular signaling will be triggered, and then elicits cytoskeleton reorganization to keep cell adhesion, cell motility and cell shape. Intergin also drives Raf/MEK/ERK pathway [35], therefore, myoblast cell to cell adhesion maybe is one trigger for the transcription of muscle-specific gene. It is reasonable that enhanced expression of these proteins in terminally differentiated myocytes strengthened cell-cell, cell-matrix adhesion and provided physical stabilization and tenacity against the tensile forces generated during muscle contraction.
Cell Structure and Motility Associated Proteins
Skeletal myogenic differentiation involved in extensive changes in cell morphology and subcellular architectures. During the differentiation process, myoblasts fuse to form multinucleated myotubes. This morphological change reflects a massive structural reorganization of cytoplasmic components including subcellular organelles [36]. Two dynamic filament systems, microtubules and microfilaments, have been considered to participate actively in generating the spatial organization of the cell [37]. Realignment of nascent α-actin and myosin into sarcomeres of myofibrils depended on microtubules network reorganization [37]. Responding to this cell-shape change, many cytoskeleton proteins are up-regulated in L6 myotubes, for instance, microtubule-associated proteins, actin related protein 2/3 complex subunit 1A (Arpc1a), transgelin, dynamin-2 and kinesin-1 etc. Microtubule-associated protein 4 is found to be required for myogensis. Antisense inhibition of muscle-specific microtubule-associated protein-4 during differentiation severely perturbed myotube formation, but had no effect on growth and cell fusion [38]. Actin-related protein 2/3 formed complex with WASP or WAVE protein to mediate the actin polymerization and the formation of branched actin networks. Transgelin, an actin cross-linking/gelling protein, is also up-regulated 4.90 fold. Dynamin-2, a microtubule-associated force-producing protein involved in producing microtubule bundles and vesicular trafficking processes [39], may be also associated with myoblasts fusion. Kinesin-1, a microtubule-dependent motor required for normal distribution of cellular components, was up-regulated by 4.4 fold, which indicates that myotube is a critical dynamic cellular component in myoblast differentiation [40]. Besides microtubules, intermediate filament is another important family of cytoskeletal proteins associated with myotubes transformation. Muscle-specific intermediate filaments (IFs) include desmin, nestin, vimentin and so on [41, 42]. These proteins are synthesized by muscle cells depends on the type of muscle and its stage of development. Desmin is presented in all muscles at all stages of development and the others appear transiently or in only certain muscles [41, 42]. In our experiment, desmin was identified but its relative expression ratio in myotubes compared to myoblast wasn’t showed by Census software. However, western blot result shows the expression of desmin was up-regulated along with myogenic process. Our SILAC data shows that nestin is highly up-regulated in Day4 L6 myotubes (7.23 fold). It is well known that nestin is a crucial component in neuron differentiation, but it is less clear how nestin functions during myogenic development.
Once myocytes form, muscle-specific contractile proteins also highly express. Skeletal muscle actin α, a basic component of thin filament, was up-regulated 2 fold. Myosins are actin-based motor proteins and the main component of thick filament. Myosin heavy chain (MHC), a myotube-specific marker, was up regulated 220 fold. Myosin heavy polypeptide 2 and myosin light polypeptide 4 also were found increased intensely in L6 myotubes comparing to myoblasts. Splice isoform 1 of tropomyosin 1 alpha and Splice Isoform 2 of tropomyosin beta increased by up to about 2 fold compared with myoblasts. Tropomyosins bind to actin filaments and in association with the troponin complex regulate muscle contraction in a calcium-dependent manner.
Taken together, all of these observations are consistent with a muscle contractility function. The up-regulation of microtubule, intermediate filament and microfilament can facilitate reframed-shape of myotubes during myogensis and maintain the structural and functional integrity of skeletal muscle.
Metabolism-related Proteins
Because skeletal muscle is force-producing contractile machinery, various metabolic events, such as ATP producing, are very active during skeletal muscle contraction. In myotubes, there is an extreme demand for ATP for muscle contraction and ATP-dependent calcium signaling. To meet this demand, skeletal muscle metabolizes large mount of glucose, fatty acids and amino acids to produce energy [43]. Consistent with which, myotubes express a large number of proteins and enhance mitochondrial function to metabolize the energy-providing products. In this study, we identified 92 proteins whose expression increased at least 1.5 fold and have been annotated to be related to energy metabolism. Among this type of proteins, 24 proteins were mapped to glucose metabolism, 19 proteins to fatty acid metabolism, 21 proteins to oxidative phosphorylation/electron transport, 13 proteins to amino acid metabolism, and 24 proteins to other metabolic functions. For example, the expression level of glyceraldehyde-3-phosphate dehydrogenase was increased by 2.76 fold, long-chain fatty acid-CoA ligase by 14.26 fold, fructose-bisphosphate aldolase A by 3.16 fold, pyruvate dehydrogenase beta by 2.91 fold, mitochondrial malate dehydrogenase by 2.04 fold, citrate synthase by 2.1 fold, all of which are consistent with Ong’s result [8]. ATP synthase B chain, ATP synthase D chain, ATP synthase beta chain and ATP synthase delta chain, the enzymes for oxidative phosphorylation, have increased significantly. GADPH, a housekeeping protein, is usually used as a control for relative protein quantification by Western blot. In this study, cells increase GADPH expression from the start of differentiation (Figure 2b), but GADPH expression was down-regulated after four days starvation (day 8) because of a decrease in glycolysis. In contrast to the Western blot results for other proteins, the decreasing expression of GADPH at day 8 suggests that GADPH may not be dominant factor in differentiation.
Transporters or Channel Proteins
Consistent with excitable and contractile characteristic of muscle, some transporter or channel proteins are highly expressed in mytubes. Skeletal muscle cells are stimulated by acetylcholine released at neuromuscular junctions by motor neurons. Ion Na+ flow into cell by Na+-K+ transporters and subsequently cells produce action potentials. Once the cells are excited, their sarcoplasmic reticulums will release through sarcoplasmic/endoplasmic reticulum calcium ATPase. Ca2+ interacts with the myofibrils and induces muscular contraction. During muscular contraction, cells consume a mass of ATP and produce substantive H+. Moreover, increase of ion Ca2+ could enhance myoblasts differentiation during the myogenic process [44]. These substrates need the help of ion transporters to pass through the cytoplasmic membranes. Ionic sodium, potassium, calcium, hydrogen transporter and cationic amino acid transporter were highly expressed in finally differentiated cell.
Among the proteins with the expression ratio of <0.5 (Table 2), some have been shown to be associated with myogenic process, such as prostaglandin F2 receptor negative regulator (PTGFRN) and prostaglandin-endoperoxide synthase 1 (COX-1). PTGFRN can inhibit the effect of PG F2 by binding to PG F2 receptor. COX-1 is a rate-control enzyme of PGs synthesis. Many studies have shown that PGs including PG F2 can promote the myogenesis by different ways [45]. If it is the case, down-regulation of PGF2 receptor negative regulator can facilitate the positive effect of PG F2 on myogenic process. But down-regulation of COX-1 looks controversial to this case. And yet, Bondesen showed recently that PG I2 can inhibit myogenesis in vitro by blocking myoblast migration and fusion [46]. So, these data indicate that there are still some controversies in myogenesis and need more detailed investigations.
4 Conclusions
In conclusion, stable isotope labeling and quantitative mass spectrometry was succeeded in analyzing skeletal-muscle differentiation. In this study, isotopic arginine was introduced in the SILAC approach, therefore, only tryptic peptides with arginine carboxyl-terminal were quantified and peptides with lysine carboxyl-terminal weren’t available for quantification. Provided that SILAC labeling with arginine and lysine would improve the accuracy of protein quantification and increase the number of peptides/proteins quantified. From our data, most of the up- or down-regulated proteins we quantified in the terminally differentiated L6 cells may provide principal or accessory support for the myogenesis process. Proteins whose expressions remained unchanged during differentiation suggest alternate mechanisms, such as modification or interactions, may be involved in muscle differentiation. For example, MAPK1 and β-catenin is the pivotal node of the signaling pathway that plays an important role in the myogenic process. Their roles are still needed further data and experiment mining. By and large, SILAC was effective in trying to elucidate the molecular mechanisms of skeletal-muscle differentiation in this study, and our data can present more clues on myogenic development. Whereas, as mentioned at the very beginning of this article, skeletal-muscle differentiation is a very complicated and dynamic process that is controlled spatio-temporally by multifarious type of factors at different transcriptional levels. The transcriptions of most proteins are dynamic, and that depends on the type of muscle, its stage of development and different species. To our viewpoint, it is essential and challenging in the future how to systematically grasp the dynamic changes of different type of proteins and their tuneful integrated functions during skeletal-muscle differentiation.
Supplementary Material
Acknowledgments
The research was supported by the National Basic Research Program of China (973) (grant no. 2004CB720004) and the National Natural Science Foundation of China (grant no. 30670587). JRY is supported by NIH P41 RR011823.
Abbreviations
- SILAC
stable isotope labeling with amino acids in cell culture
References
- 1.Wagers AJ, Conboy IM. Cellular and molecular signatures of muscle regeneration: current concepts and controversies in adult myogenesis. Cell. 2005;122:659–667. doi: 10.1016/j.cell.2005.08.021. [DOI] [PubMed] [Google Scholar]
- 2.Parker MH, Seale P, Rudnicki MA. Looking back to the embryo: defining transcriptional networks in adult myogenesis. Nat. Rev. Genet. 2003;4:497–507. doi: 10.1038/nrg1109. [DOI] [PubMed] [Google Scholar]
- 3.Lluís F, Perdiguero E, Nebreda AR, Muñoz-Cánoves P. Regulation of skeletal muscle gene expression by p38 MAP kinases. Trends. Cell. Biol. 2006;16:36–44. doi: 10.1016/j.tcb.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 4.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 5.Ong SE, Mann M. Mass spectrometry-based proteomics turns quantitative. Nat. Chem. Biol. 2005;5:252–262. doi: 10.1038/nchembio736. [DOI] [PubMed] [Google Scholar]
- 6.Tannu NS, Rao VK, Chaudhary RM, Giorgianni F, et al. Comparative proteomes of the proliferating C(2)C(12) myoblasts and fully differentiated myotubes reveal the complexity of the skeletal muscle differentiation program. Mol. Cell. Proteomics. 2004;3:1065–1082. doi: 10.1074/mcp.M400020-MCP200. [DOI] [PubMed] [Google Scholar]
- 7.Gonnet F, Bouazza B, Millot GA, Ziaei S, et al. Proteome analysis of differentiating human myoblasts by dialysis-assisted two-dimensional gel electrophoresis (DAGE) Proteomics. 2008;8:264–278. doi: 10.1002/pmic.200700261. [DOI] [PubMed] [Google Scholar]
- 8.Kislinger T, Gramolini AO, Pan Y, Rahman K, et al. Proteome dynamics during C2C12 myoblast differentiation. Mol. Cell. Proteomics. 2005;4:887–901. doi: 10.1074/mcp.M400182-MCP200. [DOI] [PubMed] [Google Scholar]
- 9.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 10.Mann M. Functional and quantitative proteomics using SILAC. Nat. Rev. Mol. Cell. Biol. 2006;7:952–958. doi: 10.1038/nrm2067. [DOI] [PubMed] [Google Scholar]
- 11.Delahunty C, Yates JR., 3rd Protein identification using 2D-LC-MS/MS. Methods. 2005;35:248–255. doi: 10.1016/j.ymeth.2004.08.016. [DOI] [PubMed] [Google Scholar]
- 12.McDonald WH, Tabb DL, Sadygov RG, MacCoss MJ, et al. MS1, MS2, and SQT-three unified, compact, and easily parsed file formats for the storage of shotgun proteomic spectra and identifications. Rapid. Commun. Mass. Spectrom. 2004;18:2162–2198. doi: 10.1002/rcm.1603. [DOI] [PubMed] [Google Scholar]
- 13.Cociorva D, Tabb DL, Yates JR., 3rd . Validation of tandem mass spectrometry database search results using DTASelect in Chapter 13:Unit 13.4 of Curr Protoc Bioinformatics. New Jersey: John Wiley & Sons Inc; 2007. [DOI] [PubMed] [Google Scholar]
- 14.Park SK, Venable JD, Xu T, Yates JR., 3rd A quantitative analysis software tool for mass spectrometry-based proteomics. Nat. Methods. 2008;5:319–322. doi: 10.1038/nmeth.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bennett AM, Tonks NK. Regulation of distinct stages of skeletal muscle differentiation by mitogen-activated protein kinases. Science. 1997;278:1288–1291. doi: 10.1126/science.278.5341.1288. [DOI] [PubMed] [Google Scholar]
- 16.Keren A, Tamir Y, Bengal E. The p38 MAPK signaling pathway: a major regulator of skeletal muscle development. Mol. Cell. Endocrinol. 2006;252:224–230. doi: 10.1016/j.mce.2006.03.017. [DOI] [PubMed] [Google Scholar]
- 17.Gredinger E, Gerber AN, Tamir Y, Tapscott SJ, Bengal E. Mitogen-activated protein kinase pathway is involved in the differentiation of muscle cells. J. Biol. Chem. 1998;273:10436–10444. doi: 10.1074/jbc.273.17.10436. [DOI] [PubMed] [Google Scholar]
- 18.Gamble SC, Chotai D, Odontiadis M, Dart DA, et al. Prohibitin, a protein downregulated by androgens, represses androgen receptor activity. Oncogene. 2007;26:1757–1768. doi: 10.1038/sj.onc.1209967. [DOI] [PubMed] [Google Scholar]
- 19.Rajalingam K, Wunder C, Brinkmann V, Churin Y, et al. Prohibitin is required for Ras-induced Raf-MEK-ERK activation and epithelial cell migration. Nat. Cell Biol. 2005;7:837–843. doi: 10.1038/ncb1283. [DOI] [PubMed] [Google Scholar]
- 20.Sun L, Liu L, Yang XJ, Wu Z. Akt binds prohibitin 2 and relieves its repression of MyoD and muscle differentiation. J. Cel. Sci. 2004;117:3021–3029. doi: 10.1242/jcs.01142. [DOI] [PubMed] [Google Scholar]
- 21.Isogai Z, Ono RN, Ushiro S, Keene DR, et al. Latent transforming growth factor beta-binding protein 1 interacts with fibrillin and is a microfibril-associated protein. J. Biol. Chem. 2003;278:2750–2757. doi: 10.1074/jbc.M209256200. [DOI] [PubMed] [Google Scholar]
- 22.Hubmacher D, Tiedemann K, Reinhardt DP. Fibrillins: from biogenesis of microfibrils to signaling functions. Curr. Top. Dev. Biol. 2006;75:93–123. doi: 10.1016/S0070-2153(06)75004-9. [DOI] [PubMed] [Google Scholar]
- 23.Gustavo A. Nader Molecular determinants of skeletal muscle mass: getting the “AKT” together. Int. J. Biochem. Cell Biol. 2005;37:1985–1996. doi: 10.1016/j.biocel.2005.02.026. [DOI] [PubMed] [Google Scholar]
- 24.Goldberg AL. Protein degradation and protection against misfolded or damaged proteins. Nature. 2003;426:895–899. doi: 10.1038/nature02263. [DOI] [PubMed] [Google Scholar]
- 25.Gardrat F, Montel V, Raymond J, Azanza JL. Proteasome and myogenesis. Mol. Biol. Rep. 1997;24:77–81. doi: 10.1023/a:1006877214153. [DOI] [PubMed] [Google Scholar]
- 26.Sun L, Trausch-Azar JS, Ciechanover A, Schwartz AL. Ubiquitin-Proteasome- mediated Degradation, Intracellular Localization, and Protein Synthesis of MyoD and Id1 during Muscle Differentiation. J. Biol. Chem. 2005;280:26448–26456. doi: 10.1074/jbc.M500373200. [DOI] [PubMed] [Google Scholar]
- 27.Srikakulam R, Winkelmann DA. Chaperone-mediated folding and assembly of myosin in striated muscle. J. Cell Sci. 2004;117:641–652. doi: 10.1242/jcs.00899. [DOI] [PubMed] [Google Scholar]
- 28.Ito H, Kamei K, Iwamoto I, Inaguma Y, Kato K. Regulation of the levels of small heat-shock proteins during differentiation of C2C12 cells. Exp. Cell. Res. 2001;266:213–221. doi: 10.1006/excr.2001.5220. [DOI] [PubMed] [Google Scholar]
- 29.Grantham J, Brackley KI, Willison KR. Substantial CCT activity is required for cell cycle progression and cytoskeletal organization in mammalian cells. Exp. Cell Res. 2006;312:2309–2324. doi: 10.1016/j.yexcr.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 30.Kamradt MC, Chen F, Sam S, Cryns VL. The small heat shock protein alpha B-crystallin negatively regulates apoptosis during myogenic differentiation by inhibiting caspase-3 activation. J. Biol. Chem. 2002;277:38731–38736. doi: 10.1074/jbc.M201770200. [DOI] [PubMed] [Google Scholar]
- 31.Covault J, Sanes JR. Distribution of N-CAM in synaptic and extrasynaptic portions of developing and adult skeletal muscle. J. Cell Biol. 1986;102:716–730. doi: 10.1083/jcb.102.3.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levi G. Cell adhesion molecules during Xenopus myogenesis. Cytotechnology. 1993;11:s91–s93. [PubMed] [Google Scholar]
- 33.Grossi A, Yadav K, Lawson MA. Mechanical stimulation increases proliferation, differentiation and protein expression in culture: Stimulation effects are substrate dependent. J. Biomech. 2007;40:3354–3362. doi: 10.1016/j.jbiomech.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 34.Brzoska E, Bello V, Darribere T, Moraczewski J. Integrin alpha3 subunit participates in myoblast adhesion and fusion in vitro. Differentiation. 2006;74:105–118. doi: 10.1111/j.1432-0436.2005.00059.x. [DOI] [PubMed] [Google Scholar]
- 35.Johnson GL, Lapadat R. Mitogen-activated protein kinase pathways mediated by ERK, JNK, and p38 protein kinases. Science. 2002;298:1911–1912. doi: 10.1126/science.1072682. [DOI] [PubMed] [Google Scholar]
- 36.Squire JM. Architecture and function in the muscle sarcomere. Curr. Opin. Struct. Biol. 1997;7:247–257. doi: 10.1016/s0959-440x(97)80033-4. [DOI] [PubMed] [Google Scholar]
- 37.Pizon V, Gerbal F, Diaz CC, Karsenti E. Microtubule-dependent transport and organization of sarcomeric myosin during skeletal muscle differentiation. EMBO J. 2005;24:3781–3792. doi: 10.1038/sj.emboj.7600842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mangan ME, Olmsted JB. Muscle-specific variant of microtubule-associated protein 4 (MAP4) is required in myogenesis. Development. 1996;122:771–781. doi: 10.1242/dev.122.3.771. [DOI] [PubMed] [Google Scholar]
- 39.Kessels MM, Dong J, Leibig W, Westermann P, Qualmann B. Complexes of syndapin II with dynamin II promote vesicle formation at the trans-Golgi network. J. Cell Sci. 2006;119:1504–1516. doi: 10.1242/jcs.02877. [DOI] [PubMed] [Google Scholar]
- 40.Ginkel LM, Wordeman L. Expression and Partial characterization of kinesin related proteins in differentiating and adult skeletal muscle. Mol. Biol. Cell. 2000;11:4143–4158. doi: 10.1091/mbc.11.12.4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vaittinen S, Lukka R, Sahlgren C, Hurme T, et al. The expression of intermediate filament protein nestin as related to vimentin and desmin in regenerating skeletal muscle. J. Neuropathol. Exp. Neurol. 2001;60:588–597. doi: 10.1093/jnen/60.6.588. [DOI] [PubMed] [Google Scholar]
- 42.Paulin D, Xue Z. Landes Bioscience and Springer US press. Austin: 2006. Desmin and Other Intermediate Filaments in Normal and Diseased Muscle in Intermediate Filaments in Intermediate Filaments; pp. 1–9. [Google Scholar]
- 43.Richardson RS. Oxygen transport and utilization: an integration of the muscle systems. Adv. Physiol. Educ. 2003;27:183–191. doi: 10.1152/advan.00038.2003. [DOI] [PubMed] [Google Scholar]
- 44.Porter GA, Makuck RF, Rivkees SA. Reduction in intracellular calcium levels inhibits myoblast differentiation. J. Biol. Chem. 2002;277:28942–28947. doi: 10.1074/jbc.M203961200. [DOI] [PubMed] [Google Scholar]
- 45.Horsley V, Pavlath GK. Prostaglandin F2 (alpha) stimulates growth of skeletal muscle cells via an NFATC2-dependent pathway. J. Cell Biol. 2003;161:111–118. doi: 10.1083/jcb.200208085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bondesen BA, Jones KA, Glasgow WC, Pavlath GK. Inhibition of myoblast migration by prostacyclin is associated with enhanced cell fusion. FASEB J. 2007;21:3338–3345. doi: 10.1096/fj.06-7070com. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.