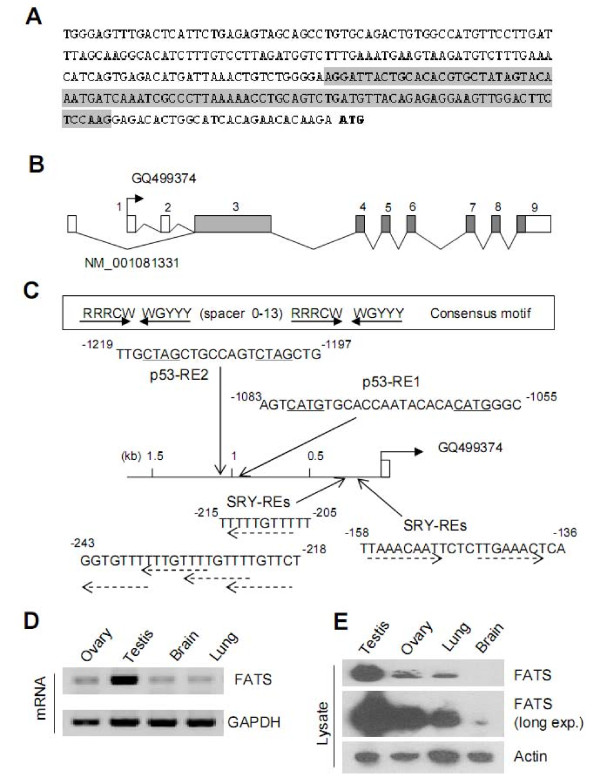
Figure 1.
Identification of a FATS transcription variant with high expression in mouse testis. (A) The sequence of 5'-UTR in FATS mRNA (GenBank accession number: GQ499374) through screening a mouse testis plasmid cDNA library (Stratagene #975304). In brief, the non-coding sequence ahead of the first coding exon of FATS gene was amplified by PCR using the primers specific to pMyr XR vector (Stratagene) and FATS coding region, respectively, and subjected to DNA sequencing subsequently. Primer sequences were as follows: pMyr-fw, 5'-ATGGGGAGTAGCAAGAGCAA-3'; FATS-rv, 5'-TCTTTTCATCAATCAGCCGG-3'. Similar approach was applied to verify the mRNA sequence downstream of the first coding exon. Primer sequences were as follows: FATS-fw, 5'-CATATTCCCGGCTGGAGTTA-3'; pMyr-rv, 5'-CTTTTCGGTTAGAGCGGATG-3'. The sequence of exon 2 is shaded, and the start codon ATG is in bold letters. (B) The genomic organization of FATS gene. FATS transcript GQ499374 differed from a previously submitted transcription variant (GenBank accession number: NM_001081331) only in 5' UTR. The coding regions of exons were shaded. (C) The p53-responsive elements (p53-RE) in mouse FATS promoter. The p53-binding site is composed of a half-site RRRCWWGYYY followed by a spacer, usually composed of 0-13 base pairs, which is then followed by a second half-site RRRCWWGYYY sequence [3,4]. R, purine; Y, pyrimidine; W, adenine or thymine. The responsive elements of the testis-determining factor SRY (SRY-REs) in mouse FATS promoter were analyzed by TFsearch program http://www.cbrc.jp/research/db/TFSEARCH.html. (D) The expression level of FATS mRNA in mouse testis and ovary was determined by RT-PCR. (E) FATS protein levels in mouse tissues. Exp.: exposure.