Abstract
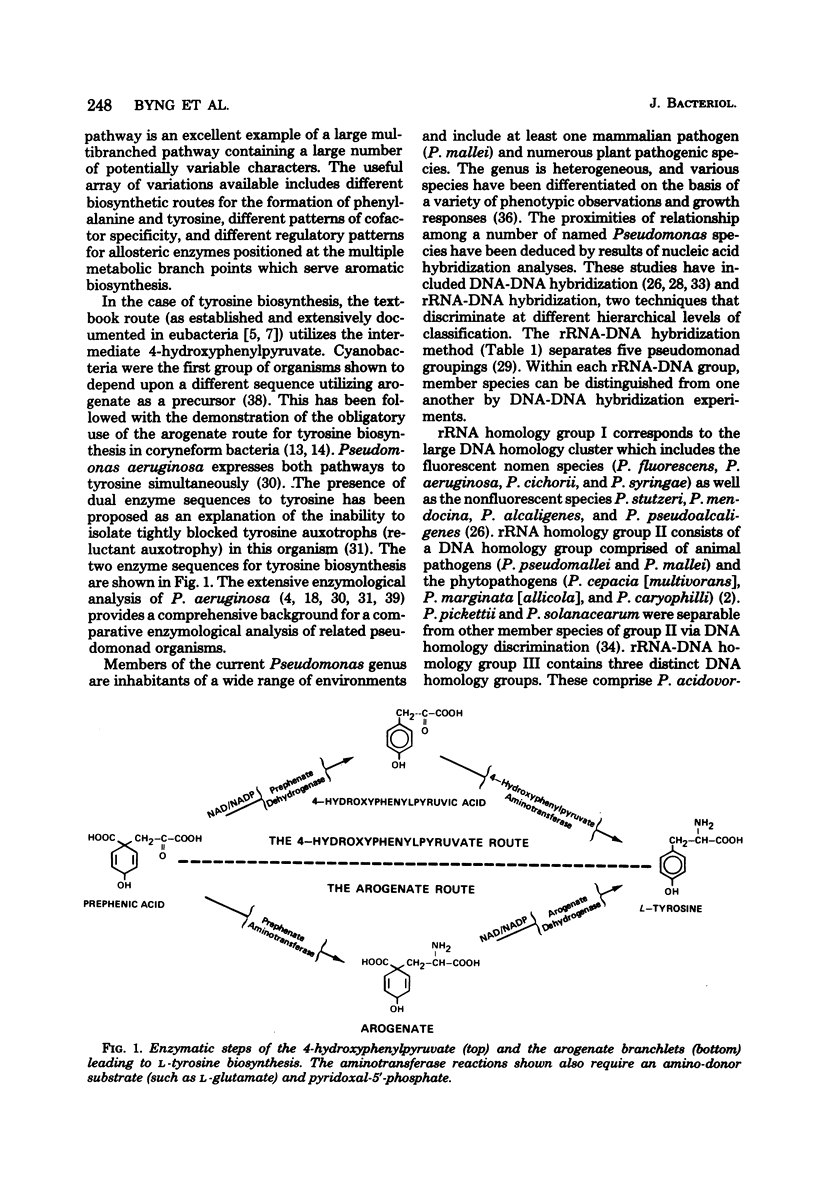
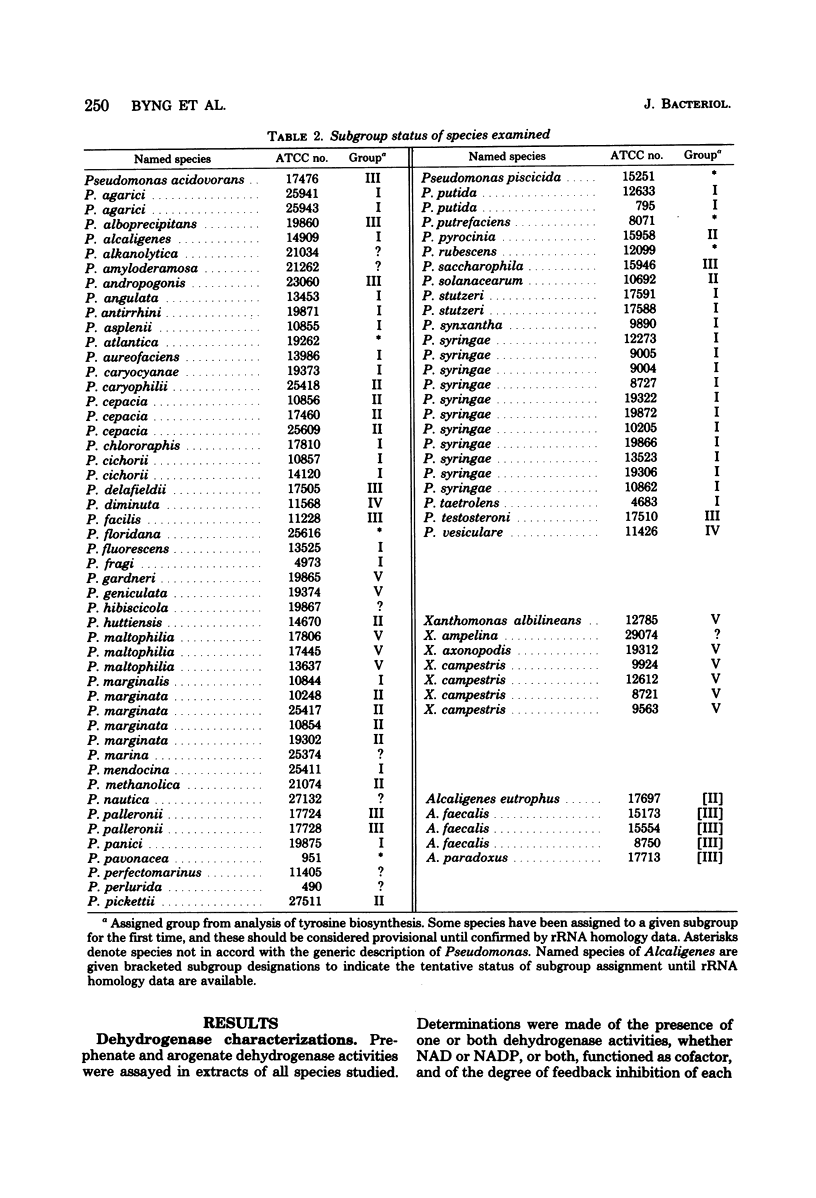
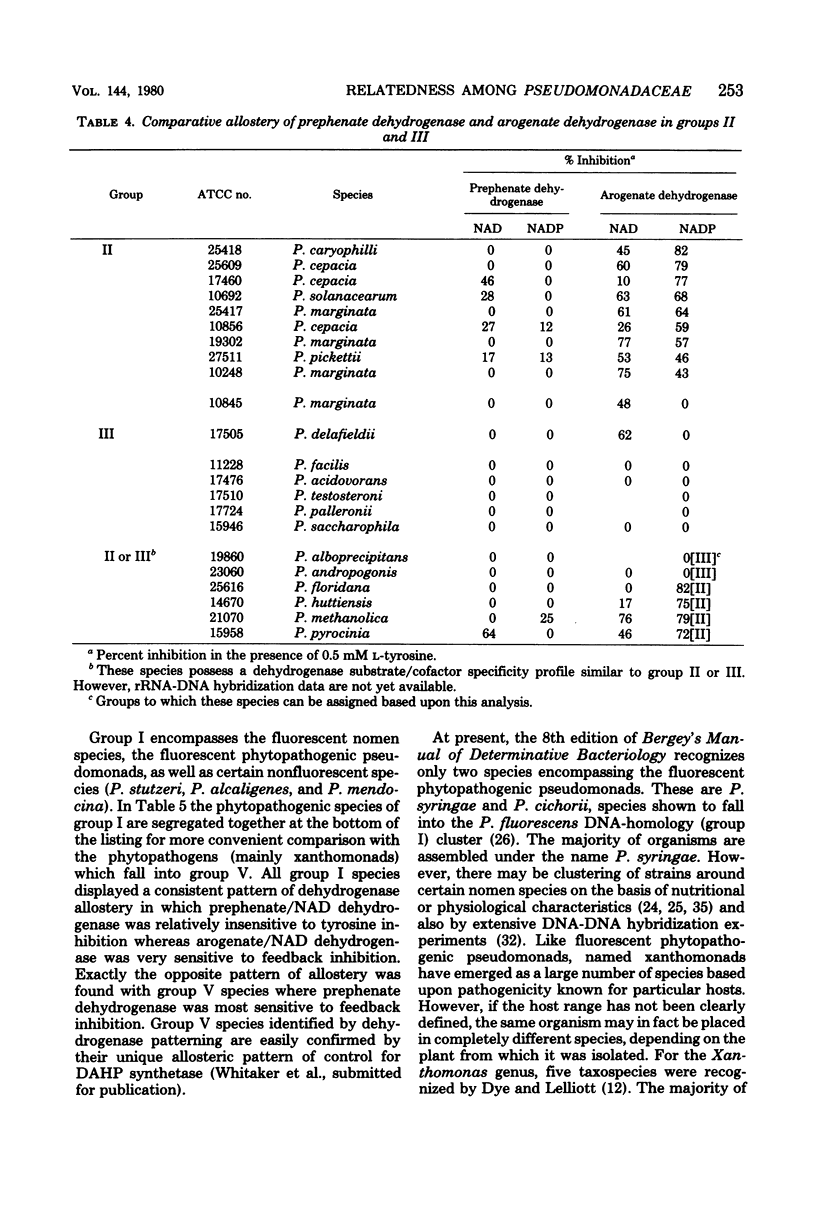
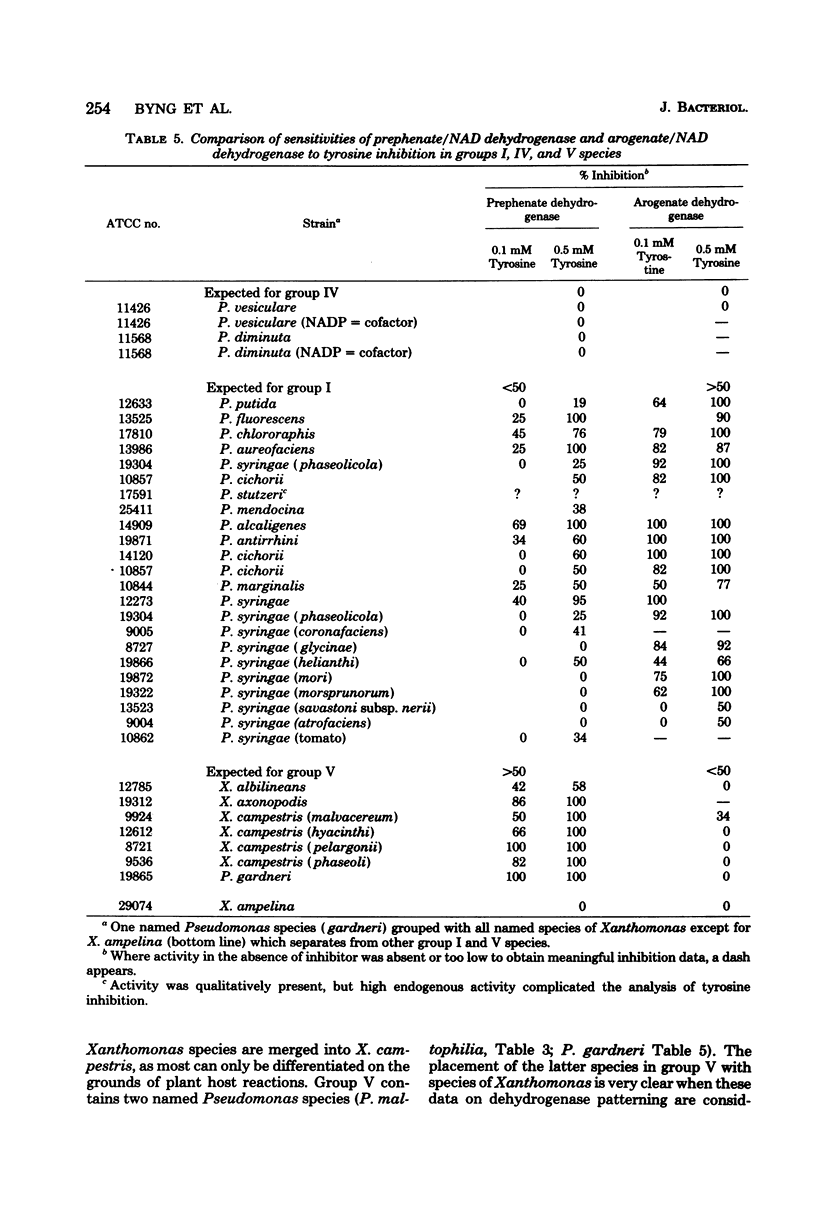
Enzymes of tyrosine biosynthesis (prephenate dehydrogenase and arogenate dehydrogenase) were characterized in 90 species currently classified within the genera Pseudomonas, Xanthomonas, and Alcaligenes. Variation in cofactor specificity and regulatory properties of the dehydrogenase proteins allowed the separation of five groups. Taxa defined by enzymological patterning corresponded strikingly with the five ribosomal ribonucleic acid (rRNA) homology groups established via rRNA-deoxyribonucleic acid hybridization. rRNA homology groups I, IV, and V all lack activity for arogenate/nicotinamide adenine dinucleotide phosphate (NADP) dehydrogenase and separated on this criterion from groups II and III, which have the activity. Group II species possess arogenate dehydrogenase enzyme (reactive with either NAD or NADP) sensitive to feedback inhibition by tyrosine, thereby separating from group III species whose corresponding enzyme was totally insensitive to feedback inhibition. The presence of prephenate/NADP dehydrogenase in group IV defined its separation from groups I and V, which lack this enzyme activity. Group I species possess an arogenate/NAD dehydrogenase that was highly sensitive to inhibition by tyrosine and a prephenate/NAD dehydrogenase of relative insensitivity to tyrosine inhibition. The opposite pattern of sensitivity/insensitivity was seen in group V species. These dehydrogenase characterizations are highly reliable for the keying of a given species to one of the five rRNA homology groups. If necessary, other confirmatory assays can be included using other aromatic pathway enzymes. These results further document the validity and utility of the approach of comparative enzymology and allostery for classification of microorganisms.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ballard R. W., Doudoroff M., Stanier R. Y., Mandel M. Taxonomy of the aerobic psuedomonads: Pseudomonas diminuta and P. vesiculare. J Gen Microbiol. 1968 Oct;53(3):349–361. doi: 10.1099/00221287-53-3-349. [DOI] [PubMed] [Google Scholar]
- Ballard R. W., Palleroni N. J., Doudoroff M., Stanier R. Y., Mandel M. Taxonomy of the aerobic pseudomonads: Pseudomonas cepacia, P. marginata, P. alliicola and P. caryophylli. J Gen Microbiol. 1970 Feb;60(2):199–214. doi: 10.1099/00221287-60-2-199. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- COTTON R. G., GIBSON F. THE BIOSYNTHESIS OF PHENYLALANINE AND TYROSINE; ENZYMES CONVERTING CHORISMIC ACID INTO PREPHENIC ACID AND THEIR RELATIONSHIPS TO PREPHENATE DEHYDRATASE AND PREPHENATE DEHYDROGENASE. Biochim Biophys Acta. 1965 Apr 12;100:76–88. doi: 10.1016/0304-4165(65)90429-0. [DOI] [PubMed] [Google Scholar]
- Calhoun D. H., Pierson D. L., Jensen R. A. Channel-shuttle mechanism for the regulation of phenylalanine and tyrosine synthesis at a metabolic branch point in Pseudomonas aeruginosa. J Bacteriol. 1973 Jan;113(1):241–251. doi: 10.1128/jb.113.1.241-251.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champney W. S., Jensen R. A. The enzymology of prephenate dehydrogenase in Bacillus subtilis. J Biol Chem. 1970 Aug 10;245(15):3763–3770. [PubMed] [Google Scholar]
- Cohen G. N., Stanier R. Y., Le Bras G. Regulation of the biosynthesis of amino acids of the aspartate family in Coliform bacteria and Pseudomonads. J Bacteriol. 1969 Sep;99(3):791–801. doi: 10.1128/jb.99.3.791-801.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis D. H., Stanier R. Y., Doudoroff M., Mandel M. Taxonomic studies on some gram negative polarly flagellated "hydrogen bacteria" and related species. Arch Mikrobiol. 1970;70(1):1–13. doi: 10.1007/BF00691056. [DOI] [PubMed] [Google Scholar]
- Fazel A. M., Bowen J. R., Jensen R. A. Arogenate (pretyrosine) is an obligatory intermediate of L-tyrosine biosynthesis: confirmation in a microbial mutant. Proc Natl Acad Sci U S A. 1980 Mar;77(3):1270–1273. doi: 10.1073/pnas.77.3.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazel A. M., Jensen R. A. Obligatory biosynthesis of L-tyrosine via the pretyrosine branchlet in coryneform bacteria. J Bacteriol. 1979 Jun;138(3):805–815. doi: 10.1128/jb.138.3.805-815.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich B., Schlegel H. G. Aromatic amino acid biosynthesis in Alcaligenes eutrophus H16. II. The isolation and characterization of mutants auxotrophic for phenylalanine and tyrosine. Arch Microbiol. 1975 Apr 7;103(2):141–149. doi: 10.1007/BF00436341. [DOI] [PubMed] [Google Scholar]
- Hill L. R. An index to deoxyribonucleic acid base compositions of bacterial species. J Gen Microbiol. 1966 Sep;44(3):419–437. doi: 10.1099/00221287-44-3-419. [DOI] [PubMed] [Google Scholar]
- Jensen R. A. Enzyme recruitment in evolution of new function. Annu Rev Microbiol. 1976;30:409–425. doi: 10.1146/annurev.mi.30.100176.002205. [DOI] [PubMed] [Google Scholar]
- Jensen R. A., Nasser D. S., Nester E. W. Comparative control of a branch-point enzyme in microorganisms. J Bacteriol. 1967 Nov;94(5):1582–1593. doi: 10.1128/jb.94.5.1582-1593.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen R. A., Zamir L., Saint Pierre M., Patel N., Pierson D. L. Isolation and preparation of pretyrosine, accumulated as a dead-end metabolite by Neurospora crassa. J Bacteriol. 1977 Dec;132(3):896–903. doi: 10.1128/jb.132.3.896-903.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelliott R. A., Billing E., Hayward A. C. A determinative scheme for the fluorescent plant pathogenic pseudomonads. J Appl Bacteriol. 1966 Dec;29(3):470–489. doi: 10.1111/j.1365-2672.1966.tb03499.x. [DOI] [PubMed] [Google Scholar]
- LéJohn H. B. Enzyme regulation, lysine pathways and cell wall structures as indicators of major lines of evolution in fungi. Nature. 1971 May 21;231(5299):164–168. doi: 10.1038/231164a0. [DOI] [PubMed] [Google Scholar]
- Misaghi I., Grogan R. G. Nutritional and biochemical comparisons of plant-pathogenic and saprophytic fluorescent pseudomonads. Phytopathology. 1969 Oct;59(10):1436–1450. [PubMed] [Google Scholar]
- Palleroni N. J., Ballard R. W., Ralston E., Doudoroff M. Deoxyribonucleic acid homologies among some Pseudomonas species. J Bacteriol. 1972 Apr;110(1):1–11. doi: 10.1128/jb.110.1.1-11.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palleroni N. J., Doudoroff M. Phenotypic characterization and deoxyribonucleic acid homologies of Pseudomonas solanacearum. J Bacteriol. 1971 Sep;107(3):690–696. doi: 10.1128/jb.107.3.690-696.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel N., Pierson D. L., Jensen R. A. Dual enzymatic routes to L-tyrosine and L-phenylalanine via pretyrosine in Pseudomonas aeruginosa. J Biol Chem. 1977 Aug 25;252(16):5839–5846. [PubMed] [Google Scholar]
- Patel N., Stenmark-Cox S. L., Jensen R. A. Enzymological basis of reluctant auxotrophy for phenylalanine and tyrosine in Pseudomonas aeruginosa. J Biol Chem. 1978 May 10;253(9):2972–2978. [PubMed] [Google Scholar]
- Ralston E., Palleroni N. J., Doudoroff M. Deoxyribonucleic acid homologies of some so-called "Hydrogenomonas" species. J Bacteriol. 1972 Jan;109(1):465–466. doi: 10.1128/jb.109.1.465-466.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sands D. C., Schroth M. N., Hildebrand D. C. Taxonomy of phytopathogenic pseudomonads. J Bacteriol. 1970 Jan;101(1):9–23. doi: 10.1128/jb.101.1.9-23.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanier R. Y., Palleroni N. J., Doudoroff M. The aerobic pseudomonads: a taxonomic study. J Gen Microbiol. 1966 May;43(2):159–271. doi: 10.1099/00221287-43-2-159. [DOI] [PubMed] [Google Scholar]
- Starr M. P., Jenkins C. L., Bussey L. B., Andrewes A. G. Chemotaxonomic significance of the xanthomonadins, novel brominated aryl-polyene pigments produced by bacteria of the genus Xanthomonas. Arch Microbiol. 1977 May 13;113(1-2):1–9. doi: 10.1007/BF00428572. [DOI] [PubMed] [Google Scholar]
- Stenmark-Cox S., Jensen R. A. Prephenate dehydrogenase from Pseudomonas aeruginosa is a regulated component of the channel-shuttle mechanism controlling tyrosine-phenylalanine synthesis. Arch Biochem Biophys. 1975 Apr;167(2):540–546. doi: 10.1016/0003-9861(75)90497-x. [DOI] [PubMed] [Google Scholar]
- Stenmark S. L., Pierson D. L., Jensen R. A., Glover G. I. Blue-green bacteria synthesise L-tyrosine by the pretyrosine pathway. Nature. 1974 Feb 1;247(5439):290–292. doi: 10.1038/247290a0. [DOI] [PubMed] [Google Scholar]


