Summary
In mammals and yeasts, oxidosqualene cyclase (OSC) catalyzes the formation of lanosterol, the first cyclic intermediate in sterol biosynthesis. We used a murine myeloma cell line (NS0), deficient in the 17β-hydroxysteroid dehydrogenase type 7 (HSD17B7), as a model to study the potential interaction of the HSD17B7 with the OSC in mammals. HSD17B7 is the orthologue of the yeast steroid-3-ketoreductase (ERG27), an enzyme of ergosterol biosynthesis that plays a protective role towards OSC. Tracer experiments with NS0 cells showed that OSC is fully active in these mammalian cells, suggesting that in mammals the ketosteroid reductase is not required for OSC activity. Mouse and human HSD17B7 were overexpressed in ERG27-deletant yeast cells, and recombinant strains were tested for (i) the ability to grow on different media, (ii) steroid-3-ketoreductase activity, and (iii) OSC activity. Recombinant strains grew more slowly than the control yeast ERG27-overexpressing strain on sterol-deficient media, whereas the growth rate was normal on media supplemented with a 3-ketoreductase substrate. The full enzymatic functionality of mammalian steroid-3-ketoreductase expressed in yeast along with the lack of (yeast) OSC activity point to an inability of the mammalian reductase to assist yeast OSC. Results demonstrate that in mammals, unlike in yeast, OSC and steroid-3-ketoreductase are non-interacting proteins.
Keywords: Cholesterol biosynthesis, NS0 cells, Saccharomyces cerevisiae, ERG7, HSD17B7, protein-protein interaction
1. Introduction
NS0 is a murine myeloma cell line that has been widely used for the production of recombinant therapeutic proteins [1,2]. These cells are cholesterol auxotrophs resulting from a transcriptional suppression of the 17β-hydroxysteroid dehydrogenase type 7 gene (HSD17B7) by DNA methylation of the promoter sequence [3,4]. 17β–Hydroxysteroid dehydrogenases are a family of enzymes involved in the synthesis and biological regulation of steroid hormones. HSD17B7 was originally reported as a prolactin receptor-associated protein catalyzing the conversion of estrone (a 17β–hydroxysteroid) to the biologically active estradiol [5]. The mammalian HSD17B7 gene was demonstrated to be the orthologue of the yeast ERG27 gene encoding the steroid-3-ketoreductase, an enzyme required for sterol biosynthesis belonging to the C4-demethylation complex [6,7]. Therefore, in mammals the enzyme HSD17B7 plays a critical role both in steroid hormone activation, by reducing 17-ketosteroid precursors, and in cholesterol biosynthesis, by reducing 3-ketosteroid intermediates produced during C-4 demethylation. Recently, A. Shehu et al. demonstrated that this enzyme plays a crucial role in mouse brain embryonic development and fetal survival, suggesting that the HSD17B7 gene belongs to genes involved in cholesterol disorders [8].
In the yeasts Saccharomyces cerevisiae [9,10], Candida albicans [11] and Candida glabrata (J. Whybrew and M. Bard, unpublished), the steroid-3-ketoreductase behaves as a moonlighting protein that, in addition to its catalytic role of reducing 3-ketosteroid intermediates in the ergosterol biosynthesis, is also involved in the functionality of oxidosqualene cyclase (OSC, Erg7p), the enzyme required for producing the first cyclical steroid intermediate of the pathway [12,13]. Yeast cells lacking the ERG27 gene, show a concomitant loss of the oxidosqualene cyclase (OSC) activity and complete ablation of sterol synthesis. Teske et al. [14] previously showed that a catalytically inactive Erg27p retains Erg7p protective function resulting in endogenous accumulation of 3-ketosteroids; however, the basis of the molecular interaction between these two enzymes remains elusive.
We used the NS0 cell line to investigate the possible relationship between steroid-3-ketoreductase and oxidosqualene cyclase in mammals. In these mammalian HSD17B7-deficient cells, the putative role of HSD17B7 as a protective agent toward oxidosqualene cyclase was evaluated using different approaches. GC-MS analysis of NS0 lipid extract and tracer experiments with intact cells indicated that OSC is fully active in these cells, suggesting that in mammals no protective action of steroid-3-ketoreductase towards OSC is necessary. The possibility of an evolutionarily attenuated interaction of mammalian 3-ketoreductases with oxidosqualene cyclase was evaluated in ERG27 deficient yeast strains overexpressing either mouse or human HSD17B7. Similar to the findings of Marijanovic et al, mammalian genes proved to complement the erg27 (3-ketoreductase) mutation poorly, resulting in hardly detectable growth after a 3 days incubation on sterol deficient media. In yeast strains overexpressing mammalian 3-ketoreductase enzymes, oxidosqualene cyclase activity was almost completely abolished. However, the mammalian genes expressed in yeast proved to be catalytically active with regard to 3-ketoreductase functionality, as shown by biochemical assays and by growth in media supplemented with sterol intermediates downstream of ERG7.
2. Materials and methods
2.1. Materials
Reagents, culture media, lanosterol, 5α-cholestan-3-one, cholesterol, ergosterol and bovine albumin were purchased from Sigma-Aldrich (Milan, Italy). Restriction enzymes and T4 DNA ligase were from Promega (Milan, Italy). Radiolabeled (R,S)-[2-14C]mevalonic acid (2.04 GBq/mmol) and [2-14C]acetic acid (2.04 GBq/mmol) were purchased from Amersham Pharmacia Biotech (UK). (3S)-[14C]-2,3-oxidosqualene was prepared as previously reported [15]. [14C]lanosterol was prepared according to the protocol described for preparing radiolabeled oxidosqualene [15], with some modifications [16]. [14C]3-ketosteroid substrate (ketosteroid 1) for the steroid-3-ketoreductase assay was prepared as described in [16]. Briefly, 0.85 mL of a yeast cell homogenate (40-50 mg of protein) prepared from an erg27 strain (SDG115) was incubated with [14C]lanosterol (0.1 mCi) in the presence of Tween 80 (0.1 mg/mL), 1 mM ATP, 1 mM NAD+ and a NADPH generating system (1 mM NADP+, 3 mM glucose-6-phosphate and 6 units of G-6-P dehydrogenase) for 16 h at 30°C with vigorous shaking. The addition of ATP and the NADPH generating system was repeated at each hour for the first 4 h. The reaction was stopped by adding 1 mL of methanolic KOH (15% w/v), and lipids were saponified at 80°C for 30 min. The nons aponifiable lipids were extracted three times with 2 mL of petroleum ether and separated on TLC plates (Kieselgel 60 F254, Merck, Darmstadt, Germany) using cyclohexane/ethyl acetate (85:15; v/v) as a developing solvent. A radioactive band with Rf 0.75 was scraped and eluted with dichloromethane. The extract was dried under nitrogen and the residue dissolved in benzene. An aliquot of the radioactive compound was treated with NaBH4 as an indicator of a 3-keto group. That the steroid scraped from the TLC plate was in fact a 3-ketosteroid was confirmed by GC-MS analysis. The scraped band was a mixture of three 3-keto-4-monomethyl steroids indistinguishable on TLC plates (4-methylzymosterone, 4-methylfecosterone and 4-methylepisterone) [16]. This mixture was designated 3-ketosteroid 1.
2.2. Mammalian cells and culture conditions
NS0 cell line was obtained from S. Buhl. Cells were routinely maintained as a suspension in either CD Hybridoma medium supplemented with 2 mM L-glutamine and 1% v/v Synthecol or Dulbecco-modified Eagle medium (DMEM) high (4500 mg/L) in glucose, supplemented with 10% fetal calf serum (FCS), 10% NCTC-109 and 1% non-essential amino acids. Cells were grown at 37°C in a humidified atmosphere (air 95%/CO2 5%).
2.3. S. cerevisiae strains and growth conditions
The genotypes of strains used in this study are listed in Table 1. The wild-type SCY876 strain was grown to early stationary phase at 30°C in YPD medium (1% yeast extract, 2% peptone, 2% glucose). BTY6-5-3 [14] and SDG115 [9] (erg27) strains were supplemented with ergosterol (0.02 mg/mL). STY35, STY36 and STY37 strains were grown in synthetic complete medium without uracil (SC-UraD: 0.17% yeast nitrogen base, 0.2% amino acids, 0.5% ammonium sulphate, 2% glucose).
Table 1.
S. cerevisiae strains used in this study.
| Strain | Genotype |
|---|---|
| SDG115 | MATa, ade5, his7, leu2-3,112, ura3-52, erg27 Δ ::URA3 |
| SCY876 | MATα, upc2-1, hap1Ty, ura3, his3, leu2, trp1 |
| BTY6-5-3 | MATα, upc2-1, hap1Ty, ura3, his3, leu2, trp1, erg27 Δ ::LEU2 |
| STY37 | MATα, upc2-1, hap1Ty, ura3, his3, leu2, trp1, erg27Δ::LEU2/pST10(ERG27) |
| STY35 | MATα, upc2-1, hap1Ty, ura3, his3, leu2, trp1, erg27Δ::LEU2/pST9 (human HSD17B7) |
| STY36 | MATα, upc2-1, hap1Ty, ura3, his3, leu2, trp1, erg27Δ::LEU2/pST7 (mouse HSD17B7) |
For spot plate analyses, strains were grown on SC-UraD supplemented with either lanosterol, ergosta-7,22-diene-3-one (precursor to Erg27p), ergosterol or Tween 80 (control) and grown for 3 d.
2.4. Cloning and expression of recombinants human and mouse steroid-3-ketoreductase in S. cerevisiae BTY6-5-3
Plasmids pGEX-human HSD17B7 and pGEX-mouse HSD17B7 were obtained from J. Adamski. These plasmids were digested, using BamHI and HIndIII for human HSD17B7 and BamHI and KpnI for mouse HSD17B7, and the resultant fragments were cloned into p426GPD yeast plasmid vector (Mumberg et al., 1995) under the control of GPD promoter to yield the pST9 and pST7 plasmids.
The yeast control ERG27 gene was cloned from pST3 plasmid (p425GPD-ERG27) by digesting the vector with SpeI and SalI. The resultant fragment was cloned into p426GPD vector to yield the pST10 plasmid.
All plasmids were maintained in Escherichia coli DH5α cultured in LB medium supplemented with ampicillin (100 μg/mL). Plasmid DNA was sequenced by the C.R.I.B.I.-BMR Servizio Sequenziamento DNA, Padova (Italy).
Lithium acetate was used to transform the S. cerevisiae steroid-3-ketoreductase mutant strain, BTY6-5-3, with the pST9, pST7 and pST10 plasmids [17] to generate the yeast strains STY35, STY36 and STY37, respectively. Transformants were selected on synthetic complete medium plates without uracil (SC-UraD) supplemented with ergosterol (0.02 mg/mL).
2.5. Incorporation of [2-14C] acetate into nonsaponifiable lipids of NS0 cells
Sterol biosynthesis in NS0 cells was followed by incorporation of [2-14C]acetate into nonsaponifiable lipids. Cells were seeded in 24-well plastic clusters. Each well received 1×105 cells suspended in 1 mL of either Dulbecco-modified Eagle medium (DMEM) containing 10% lipid-depleted serum [18] or CD Hybridoma medium. After either a 2 or 24 h incubation, [2-14C]acetate (1 μCi/well) was added and cells incubated for a further 3 h. The medium was then removed and cells were treated with 0.1 N NaOH (500 μL) for 30 min. Phosphate buffer-saline (PBS) (500 μL) was then added to each cell lysate, and the mixture was saponified by treatment with methanolic KOH (1 mL, 10% w/v) for 30 min at 80°C. Nonsaponifiable lipids were extra cted twice with 1.5 mL of petroleum ether, and an aliquot of the extracts was incubated with 5 mg of NaBH4 in 1 mL of ethanol for 30 min with magnetic stirring. This reaction was stopped by adding 1 mL of water, and each reaction mixture was then extracted twice with 1 mL of petroleum ether. NaBH4 −treated and −non treated extracts were separated on TLC plates (Alufolien Kieselgel 60 F254, Merck, Darmstadt, Germany) using cyclohexane/ethyl acetate (85:15 v/v) as a developing solvent. Authentic standards of cholesterol, lanosterol, 5α-cholestan-3-one, dioxidosqualene, oxidosqualene and squalene were added on TLC plates as references. Radioactivity in separated bands was measured using a System 200 Imaging Scanner (Hewlett-Packard, Palo Alto, CA, USA).
2.6. Incorporation of [2-14C]acetate into nonsaponifiable lipids of S. cerevisiae cells
Sterol biosynthesis in whole yeast cells was followed by incorporation of [2-14C]acetate into nonsaponifiable lipids as previously described [19]. Briefly, washed cells (1×108 cells) were resuspended in 1 mL of fresh media, incubated with 0.1 μCi of [2-14C]acetate and shaken for 3 h at 30°C. Then, cells were saponified in 1 mL of methanolic KOH (15% w/v) for 30 min at 80°C. Nonsaponifiable lipids were extracted twice with 1.5 mL of petroleum ether and separated on TLC plates (Alufolien Kieselgel 60 F254, Merck, Darmstadt, Germany) using cyclohexane/ethyl acetate (85:15; v/v) as a developing solvent. Authentic standards of ergosterol, lanosterol, 3-ketosteroid 1, dioxidosqualene, oxidosqualene and squalene were added on TLC plates as references. Radioactivity in separated bands was measured using a System 200 Imaging Scanner (Hewlett-Packard, Palo Alto, CA, USA).
2.7. Erg7 and Erg27 enzyme assays with radiolabeled substrates
Cell-free homogenates were prepared as described [20]. Briefly, after lysis of the yeast cell wall with lyticase, the spheroplasts were homogenized with a Potter device. Proteins in the homogenate were quantified using the SIGMA Protein Assay Kit [21], with bovine serum albumin as a standard.
The enzyme assay for Erg7p has been described previously [20]. Cell homogenates (1.5 mg of protein) were incubated for 1 h at 35°C with (3S)-[14C]-2,3-oxidosqualene (4.5×10−4 mCi) and the enzymatic reaction resulting in lanosterol synthesis was terminated by adding 1 mL of methanolic KOH (15% w/v). Lipids were saponified at 80°C for 30 min and nonsaponifiable l ipids were extracted with petroleum ether. Extracts were then spotted on TLC plates (Alufolien Kieselgel 60 F254, Merck, Darmstadt, Germany) and developed in cyclohexane/ethyl acetate (85:15; v/v). The conversion of labeled oxidosqualene to labeled lanosterol was quantified with a System 200 Imaging Scanner (Hewlett-Packard, Palo Alto, CA, USA).
The enzyme assay for Erg27p has been described [16]. Briefly, cell homogenates (3 mg of protein) were incubated for 3 h at 30°C with [ 14C]3-ketosteroid 1 (7×10−4 μCi), Tween 80 (0.1 mg/mL), 1 mM ATP and a NADPH generating system (indicated above) in 0.1 M Tris (pH 7.4, final volume 0.5 mL) buffer containing 5 mM MgCl2 and 2 mM MnCl2,. The reaction was terminated by adding 1 mL of methanolic KOH (15% w/v), and lipids were saponified at 80°C for 30 min. The nonsaponifiable lipids were extracted twice with 1.5 mL of petroleum ether, spotted on TLC plates and developed in cyclohexane/ethyl acetate (85:15; v/v). The conversion of the labeled 3-ketosteroid 1 to labeled reaction products was quantified with a System 200 Imaging Scanner (Hewlett-Packard, Palo Alto, CA, USA). Enzyme activity was expressed as a ratio of products formed/starting compound. The actual enzyme specific activity (amount of product formed/time/mg protein) could not be calculated as the specific radioactivity of the radiolabeled substrates could not be determined.
2.8. GC analysis of the nonsaponifiable lipids extracted from NS0 and yeast cells
NS0 cells were grown in CD Hybridoma medium + 1% Synthecol. STY35, STY36 and STY37 yeast strains were grown overnight in SC-UD media supplemented with cholesterol (0.02 mg/mL). NS0 and yeast cells were then harvested and saponified for 1 h at 80°C in ethanolic KOH (25% w/v). The nonsaponifiab le lipids were extracted three times with one volume of n-heptane and analyzed by GC-MS. Gas-chromatographic-mass spectrometric analyses to determine sterol profiles were carried out as previously described [22].
3. Results
3.1. Sterol accumulation profile in NS0 cells
In both yeast and mammalian cells, grown aerobically in the presence of radiolabeled acetate, a large amount of ergosterol/cholesterol accumulates, whereas a small amount of oxidosqualene and a negligible amount of dioxidosqualene forms [15,23,24]. The accumulation of oxidosqualenes (mainly dioxidosqualene), is indicative of reduced oxidosqualene cyclase functionality [19,25]. To test the functionality of oxidosqualene cyclase in the NS0 cell line, pulse labelling experiments were performed. NS0 cells were pre-cultured for 2 or 24 h in lipid-depleted media to stimulate the uptake of radiolabeled acetate during a 3 h pulse period at 37°C. Following the pulse, nonsaponifiable lipids were extracted and separated on TLC plates along with authentic standards of squalene, oxidosqualene, dioxidosqualene, lanosterol (as a reference for 4,4-dimethylsterols), 5α-cholestan-3-one (same Rf of dioxidosqualene) and cholesterol (as a reference for 4,4-desmethylsterols). Results are reported in Table 2. In all the extracts from cells cultured under different conditions, a radioactive peak with a Rf value between oxido- and dioxidosqualene (Rf 0.75) was detected. This peak was identified as a keto compound after treating an aliquot of the lipid extract with NaBH4, a reagent that effectively reduces the keto groups to the corresponding alcohols [16]. In the treated extracts separated by TLC, the peak with Rf 0.75 was no longer present, whereas two more polar compounds, presumably the corresponding 3-α and 3-β hydroxy derivatives, were observed. Comparison of the chromatographic behaviour of this keto compound with ketosteroid 1 formed by erg27 yeast strains (a mixture of 4-methylzymosterone, 4-methylfecosterone and 4-methylepisterone [16]) suggested it was a 4-monomethyl sterone. The accumulation of a radiolabeled 3-ketosteroid as well as the negligible presence of oxidosqualenes suggest that OSC activity is normal in NS0 cells and that the presence of steroid 3-ketoreductase does not appear to be necessary for OSC activity in mammalian cells. Presence of a small amount of desmethylsterols (Table 2) may be explained by considering that in NS0 cells 17b-hydroxysteroid dehydrogenase type 7 gene (HSD17B7) is epigenetically silenced, not disrupted: therefore, a very low expression of active enzyme could occur [4].
Table 2.
TLC separation of nonsaponifiable lipid fraction of NS0 cells incubated with [14C]acetate.
| Culture conditions |
% Total radioactivity incorporated |
|||||
|---|---|---|---|---|---|---|
| Squalene | Oxido squalene |
4-mono methyl- sterone |
Dioxidosqualene/ 5α-cholestan-3-one* |
4,4- dimethyl- sterols |
4,4- desmethyl- sterols |
|
| a | 4.95 | 3.64 | 51.19 | 2.62 | 23.90 | 13.70 |
| b | 1.11 | 2.21 | 57.43 | 2.73 | 29.15 | 7.37 |
| c | 5.63 | 4.51 | 58.57 | 3.17 | 15.44 | 12.68 |
| d | 4.64 | 2.29 | 61.18 | 4.48 | 23.65 | 3.76 |
Dioxidosqualene and 5α-cholestan-3-one were indistinguishable on TLC.
NS0 cells were pre-cultured for 2 h in the Dulbecco-modified Eagle medium (DMEM) containing 10% (v/v) lipid-depleted serum.
NS0 cells were pre-cultured for 24 h in the Dulbecco-modified Eagle medium (DMEM) containing 10% (v/v) lipid-depleted serum.
NS0 cells were pre-cultured for 2 h in the CD Hybridoma medium.
NS0 cells were pre-cultured for 24 h in the CD Hybridoma medium.
Results are the means of two separated experiments with duplicate incubations, each. The maximum deviations from the mean were less than 10%.
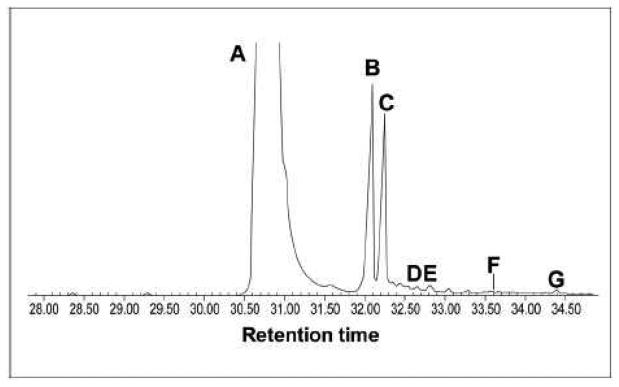
The accumulation of a ketosterone-type compound was confirmed by GC-MS analysis of nonsaponifiable lipids extracted from NS0 cells routinely maintained in a cholesterol-enriched medium. Besides cholesterol added exogenously to support growth, and its degradation product cholesta-4-en-3-one (often observed when cholesterol is added to the growth medium), GC-MS analysis of the lipid extract revealed the presence of a major peak corresponding to 4-methylcholesta-8,24-dien-3-one (4-methylzymosterone) as well as smaller amounts of other 4-methylcholesta-3-ones (Fig. 1). These data further support that a 3-ketoreductase deficiency in mammals leads to accumulation of 3-ketosteroids rather than oxidosqualenes and demonstrate that the interaction between OSC and 3-ketoreductase is not conserved from yeast to mammals.
Fig. 1.
GC-MS profile of nonsaponifiable lipid extract from NS0 cells grown in the presence of cholesterol: A, cholesterol; B, cholesta-4-en-3-one; C, 4-methylcholesta-8,24-dien-3-one; D, 4-methylcholesta-7,24-dien-3-one; E, 4-methylcholesta-dien-3-one + m/z 398; F, lanosterol + 4,4-dimethylzymosterol; G, lanosta-trien-3-one. NS0 cells accumulate 3-ketosteroid compounds and not oxidosqualenes.
3.2. Analyses of erg27 yeast strains expressing the yeast, human and mouse steroid-3-ketoreductase
Marijanovic et al. demonstrated that HSD17B7 is the mammalian steroid-3-ketoreductase by complementation of an S. cerevisiae erg27 (3-ketoreductase) strain [7]. To further investigate the complementation properties of various mammalian 3-ketoreductases, we constructed three different yeast strains overexpressing in an erg27 background respectively human, mouse or yeast 3-ketoreductase. Yeast transformant colonies were then tested for (i) growth on media with and without various sterols, (ii) 3-ketoreductase catalytic activity and (iii) oxidosqualene cyclase activity.
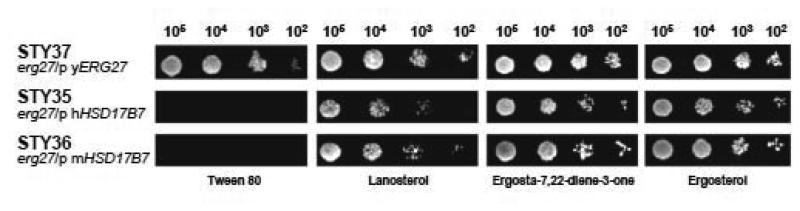
Yeast transformants containing human, mouse or yeast 3-ketoreductase genes were grown on either sterol-depleted medium or media supplemented with various sterols or sterol precursors. Consistent with the findings of Marijanovic, spot plate analysis (Fig. 2) clearly indicated that both human and mouse 3-ketoreductase complement yeast very poorly in the absence of exogenous sterol. However, when the medium was supplemented with lanosterol, the first sterol intermediate and the product of the oxidosqualene cyclase reaction, the growth rate was improved. Further, when media was supplemented with ergosta-7,22-diene-3-one, a substrate of Erg27p, the growth was comparable to that obtained in media containing ergosterol, suggesting a full catalytic functionality of the mammalian HSD17B7 enzyme in the yeast erg27 background (Fig. 2).
Fig. 2.
Spot plate analysis of erg27 mutant yeast strains transformed with p426GPD containing the human, mouse, or yeast 3-ketoreductase. Strains were grown on SC-Ura media containing glucose supplemented with either lanosterol, ergosta-7,22-diene-3-one (precursor to Erg27p), ergosterol or Tween 80 (control). Serial dilutions of 105-102 cells were spotted onto indicated plates. Pictures were taken after 3 days growth. Strains expressing mammalian steroid-3-ketoreductase complement very poorly the erg27 yeast strain in the absence of exogenous sterol; however they complement well in the presence of ergosta-7,22-diene-3-one.
The enzymatic properties of yeast and mammalian 3-ketoreductases expressed in yeast were also assessed by direct determination of the enzymatic activity of cell homogenates incubated with a radiolabeled 3-ketosteroid (see Materials and methods). In both yeast and mammalian 3-ketoreductase-expressing strains, the substrate was transformed into the corresponding 3-hydroxyderivative, which partially was converted to the end product ergosterol (Table 3). The steroid-3-hydroxyderivative was identified by comparing the products of enzymatic and chemical (NaBH4) reduction of the 3-keto-substrate (data not shown) [16]. In strains expressing mammalian 3-ketoreductase, the transformation of the 3-ketosubstrate (>50%) to the hydroxy derivative was comparable to that observed in wt and yeast ERG27-overexpressing strains (Table 3).
Table 3.
Incubation of homogenates from yeast strains with [14C]3-ketosteroid 1.
| Strain | % Radioactivity in nonsaponifiable lipids |
||
|---|---|---|---|
| 3-Ketosteroid 1 | 3-Hydroxysteroid 1 | Ergosterol | |
| SCY876 (wt) | 42.14 | 30.62 | 27.24 |
| BTY6-5-3 (erg27) | 92.02 | 7.98 | ND |
| STY37 (erg27/p yERG27) | 31.89 | 50.73 | 17.38 |
| STY35 (erg27/p hHSD17B7) | 39.17 | 41.57 | 19.26 |
| STY36 (erg27/p mHSD17B7) | 48.05 | 49.27 | 2.68 |
Ketosteroid 1 is a mixuture of 4-methylzymosterone, 4-methylfecosterone and 4-methylepisterone.
Results are the means of two separated experiments with duplicate incubations, each. The maximum deviations from the mean were less than 10%; ND, not detectable.
Oxidosqualene cyclase activity of strains was tested both by incubating growing cells with radiolabeled acetate and by incubating cell homogenates with radiolabeled oxidosqualene, the substrate of the enzyme. Experiments with growing cells showed a clear difference between yeast and mammalian 3-ketoreductase-expressing strains. In both yeast wt and yeast ERG27-overexpressing strains, radioactivity accumulated mainly into ergosterol, the end product of the pathway, whereas in both mouse and human 3-ketoreductase-expressing strains more than 80% radioactivity accumulated into epoxysqualenes (mono- and dioxide) (Table 4), a result expected if the oxidosqualene cyclase is almost totally inactive. Similar results have been obtained in other studies (i) with strains lacking the Erg7p (oxidosqualene cyclase) [15], (ii) with yeast strains lacking the Erg27p (yeast 3-ketoreductase), the “protector” of yeast oxidosqualene cyclase [10], or when yeast cells are treated with an inhibitor of oxidosqualene cyclase [19,25].
Table 4.
TLC separation of nonsaponifiable lipid fraction of S. cerevisiae cells incubated with [14C]acetate.
| Strain | % Total radioactivity incorporated |
|||||
|---|---|---|---|---|---|---|
| Squalene | Oxido squalene |
3-Keto steroid 1 |
Dioxido squalene |
Lanosterol | Ergosterol | |
| SCY876 (wt) | 14.09 | 4.58 | 3.60 | 1.63 | 8.52 | 67.58 |
| BTY6-5-3 (erg27) | 5.38 | 14.43 | 4.04 | 75.25 | 0.41 | 0.49 |
| STY37 (erg27/p yERG27) | 15.14 | 3.72 | 6.07 | 4.28 | 5.26 | 65.53 |
| STY35 (erg27/p hHSD17B7) | 2.78 | 28.93 | 3.39 | 51.94 | 3.18 | 9.78 |
| STY36 (erg27/p mHSD17B7) | 2.04 | 36.56 | 1.76 | 56.69 | 1.13 | 1.82 |
Ketosteroid 1 is a mixture of 4-methylzymosterone, 4-methylfecosterone and 4-methylepisterone.
Results are the means of two separated experiments with duplicate incubations, each. The maximum deviations from the mean were less than 10%.
Experiments with cell homogenates incubated with radiolabeled oxidosqualene, in which Erg7p specific activities were assayed, confirmed these results. In cell homogenates prepared from wt and yeast ERG27-overexpressing strains, radiolabeled oxidosqualene was effectively transformed into lanosterol, whereas in homogenates from mammalian-3-ketoreductase-expressing strains, oxidosqualene cyclase activity was negligible (Table 5).
Table 5.
Oxidosqualene cyclase specific activity in homogenates from yeast strains.
| Strain | OSC specific activity (nmol lanosterol/h/mg protein) |
|---|---|
| SCY876 (wt) | 0.91 |
| BTY6-5-3 (erg27) | 0.05 |
| STY37 (erg27/p yERG27) | 0.77 |
| STY35 (erg27/p hHSD17B7) | 0.02 |
| STY36 (erg27/p mHSD17B7) | 0.04 |
Results are the means of two separated experiments with duplicate incubations, each. The maximum deviations from the mean were less than 10%.
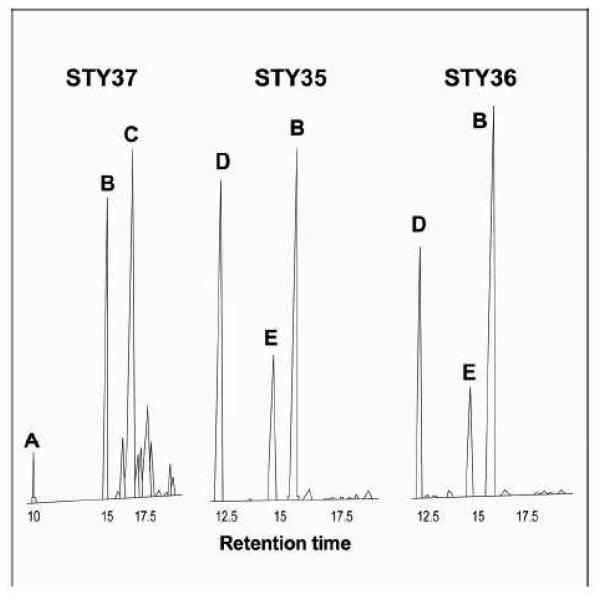
GC-MS analyses confirmed that yeast cells expressing either mouse or human HSD17B7, grown in a cholesterol supplemented medium, accumulate squalene epoxides (Fig. 3).
Fig. 3.
GC-MS profiles of nonsaponifiable lipid extracts from yeast cells grown in the presence of cholesterol: A, squalene; B, cholesterol; C, ergosterol; D, oxidosqualene; E, dioxidosqualene. The control strain, STY37 (erg27/p yERG27), accumulates ergosterol, whereas the strains expressing mammalian steroid-3-ketoreductase, STY35 (erg27/p hHSD17B7) and STY36 (erg27/p mHSD17B7), accumulate oxidosqualenes.
4. Discussion
The present study highlights differences between yeast Saccharomyces cerevisiae and mammals regarding the interaction of the steroid-3-ketoreductase with oxidosqualene cyclase, two essential enzymes of sterol biosynthesis. In the yeast Saccharomyces cerevisiae [10] (as well as in Candida albicans [11] and Candida glabrata [J. Whybrew and M. Bard, unpublished]), the absence of the 3-ketoreductase completely abolishes the functionality of oxidosqualene cyclase. By studying sterol biosynthesis in NS0 cells, a mouse cell line auxotrophic for cholesterol, specifically deficient in 3-ketoreductase activity [3], we observed that in these mammalian cells oxidosqualene cyclase is active. Both tracer experiments and lipid analysis led to this conclusion. In cells incubated with radiolabeled acetate, no oxido- or dioxidosqualene (indicative of low or absent oxidosqualene cyclase activity) accumulated, whereas a 3-keto-4-monomethylsteroid accumulated at significant levels (50 to over 60% in the nonsaponifiable extracts, Table 2). A ketosteroid compound was also the main component that accumulated in the GC-MS analyzed nonsaponifiable fraction extracted from NS0 cells (Fig. 1). Overall, these results suggest that the OSC is active in NS0 cells, thus the protective role of the 3-ketoreductase with respect to oxidosqualene cyclase activity is not conserved in mammals. This conclusion poses an intriguing question regarding the divergent evolution of the steroid-3-ketoreductase enzymes in yeast and mammals. Marijanovic et al. [7] reported that both human and mouse 3-ketoreductase genes complemented the yeast ERG27 deletion and found that transformants expressing the mammalian enzymes grew much more slowly than a strain expressing the yeast ERG27 gene. We repeated these complementation experiments with new yeast recombinant strains overexpressing mammalian 3-ketoreductases (lacking, in our case, GST reporter group) and confirmed that the yeast ERG27 gene complements much better than mouse or human gene. Marijanovic et al. suggested that the slower growth was due to low expression of the mammalian enzyme in a yeast system. However, our data prove that the mammalian enzymes are catalytically active in yeast and complement the enzymatic function of Erg27p, allowing normal growth in media supplemented with lanosterol or 3-ketosteroids. Conversely, OSC is inactive in the yeast strains transformed with the human or mouse 3-ketoreductases, resulting in a severe growth defect. Therefore, in these recombinant strains, the poor protection of oxidosqualene cyclase rather than the low expression of mammalian proteins could explain the slow growth.
The failure of mammalian 3-ketoreductase to fulfill the role of protecting Erg7p in yeast suggests that in passing from yeasts to mammals these enzymes maintained the catalytic function, but lost the chaperone-like properties. What may account for the acquired independence of mammalian oxidosqualene cyclase from 3-ketoreductase? Comparison of the catalytic properties of yeast Erg27p and mammalian HSD17B7 enzymes allows speculation about the evolution of the 3-ketoreductase. The mammalian enzyme has previously been described as catalyzing also the conversion of steroid estrone to estradiol, thus playing a key role both in cholesterol synthesis and in steroid hormone synthesis. Breitling et al. [6] suggested that mammalian enzyme (HSD17B7) is derived from proteins that probably reduced zymosterone or a similar compound to the corresponding alcohols and has only recently acquired its additional function in estradiol synthesis. This acquisition might have been accomplished by the loss of other functions such as the protective function towards the OSC. To support this speculation, additional experiments with novel yeast recombinant strains are necessary. For example, we do not know if the mammalian oxidosqualene cyclase maintains its independence when expressed in an erg27 yeast strain. If not, it would mean that the yeast background strongly influences the interaction between oxidosqualene cyclase and 3-ketoreductase.
In this case, it would be interesting to establish which is the best protector of mammalian cyclase expressed in yeast: whether the yeast 3-ketoreductase, which is closer to the cellular background, or the mammalian 3-ketoreductase, which is phylogenetically closer to mammalian oxidosqualene cyclase.
Likely, a more conclusive explanation of the evolutionary relationship between the OSC cyclase and 3-ketoreductase will come from extending the gene disruption approach to a wider series of organisms. Among these, two species may be particularly interesting: the yeast Schizosaccharomyces pombe and the plant pathogenic fungus Botrytys cinerea. S. pombe shares some of the features of mammalian cells with regard to sterol synthesis such as the finding that the sterol C-4 demethylase complex is involved in the adaptation to low oxygen [26,27]. B. cinerea is responsible for grey mold diseases and its 3-ketoreductase has been recently recognized as the specific target of fenhexamid, one of the latest antibotrytis fungicides [28,29]: would the inhibition of 3-ketoreductase affect OSC activity? Finally, the absence of interaction between mammalian 3-ketoreductase (HSD17B7) and oxidosqualene cyclase suggests that it is worth considering the protein interaction in fungal cells as a potential target for the development of specific antifungal drugs able to disrupt the yeast interaction even if they didn't inhibit 3-ketoreductase itself.
Research Highlights.
In NS0 cells, unlike in yeast, 3-ketoreductase does not assist oxidosqualene cyclase
Mouse and human 3-ketoreductases are catalytically active in yeast
Mouse and human 3-ketoreductases don't assist yeast oxidosqualene cyclase
When, from yeasts to mammals, the reductase- cyclase interaction, was lost?
Acknowledgments
We thank Prof. J. Adamski (GSF-National Research Center for Environment and Health, Institute of Experimental Genetics, Genome Analysis Center, Neuherberg, Germany) for providing plasmids pGEX-human HSD17B7 and pGEX-mouse HSD17B7, and Susan Buhl (Department of Cell Biology, Albert Einstein College of Medicine, NY, USA) for providing NS0 cells. This investigation was supported by the regional government (Regione Piemonte Ricerca Sanitaria Finalizzata 2009) and the Ministero dell'Istruzione, Università e Ricerca (MIUR) (to G. B.), and by National Institutes of Health (Grant GM62104 to M. B.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Zhou WC, Chen C-C, Buckland B, Aunins JG. Fed-batch culture of recombinant NS0 myeloma cells with high monoclonal antibody production. Biotechnol. Bioeng. 1997;55:783–792. doi: 10.1002/(SICI)1097-0290(19970905)55:5<783::AID-BIT8>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 2.Chu L, Robinson DK. Industrial choice for protein production by large scale cell cultures. Curr. Opin. Biotechnol. 2001;12:180–187. doi: 10.1016/s0958-1669(00)00197-x. [DOI] [PubMed] [Google Scholar]
- 3.Seth G, McIvor RS, Hu W-S. 17b-Hydroxysteroid dehydrogenase type 7 (Hsd17b7) reverts cholesterol auxotrophy in NS0 cells. J. Biotechnol. 2006;121:241–252. doi: 10.1016/j.jbiotec.2005.07.017. [DOI] [PubMed] [Google Scholar]
- 4.Seth G, Ozturk M, Hu W-S. Reverting cholesterol auxotrophy of NS0 cells by altering epigenetic gene silencing. Biotechnol. Bioeng. 2006;93:820–827. doi: 10.1002/bit.20720. [DOI] [PubMed] [Google Scholar]
- 5.Nokelainen P, Peltoketo H, Vihko R, Vihko P. Expression cloning of a novel estrogenic mouse 17 b-hydroxysteroid dehydrogenase/17-ketosteroid reductase (m17HSD7), previously described as a prolactin receptor-associated protein (PRAP) in rat. Mol. Endocrinol. 1998;12:1048–1059. doi: 10.1210/mend.12.7.0134. [DOI] [PubMed] [Google Scholar]
- 6.Breitling R, Krazeisen A, Möller G, Adamski J. 17b-hydroxysteroid dehydrogenase type 7—an ancient 3-ketosteroid reductase of cholesterogenesis. Mol. Cell. Endocrinol. 2001;171:199–204. doi: 10.1016/s0303-7207(00)00416-0. [DOI] [PubMed] [Google Scholar]
- 7.Marijanovic Z, Laubner D, Möller G, Gege C, Hunsen B, Adamski J, Breitling R. Closing the gap: identification of human 3-ketosteroid reductase, the last unknown enzyme of mammalian cholesterol biosynthesis. Mol. Endocrinol. 2003;17:1715–1725. doi: 10.1210/me.2002-0436. [DOI] [PubMed] [Google Scholar]
- 8.Shehu A, Mao J, Gibori GB, Halperin J, Le J, Devi YS, Merrill B, Kiyokawa H, Gibori G. Prolactin Receptor-Associated Protein/17β-Hydroxysteroid Dehydrogenase Type 7 Gene (Hsd17b7) Plays a Crucial Role in Embryonic Development and Fetal Survival. Mol. Endocrinol. 2008;22:2268–2277. doi: 10.1210/me.2008-0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gachotte D, Sen SE, Eckstein J, Barbuch R, Krieger M, Ray BD, Bard M. Characterization of the Saccharomyces cerevisiae ERG27 gene encoding the 3-keto reductase involved in C-4 sterol demethylation. Proc. Natl. Acad. Sci. 1999;96:12655–12660. doi: 10.1073/pnas.96.22.12655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mo C, Milla P, Athenstaedt K, Ott R, Balliano G, Daum G, Bard M. In yeast sterol biosynthesis the 3-ketoreductase protein (Erg27p) is required for oxidosqualene cyclase (Erg7p) activity. Biochim. Biophys. Acta. 2003;1633:68–74. doi: 10.1016/s1388-1981(03)00088-x. [DOI] [PubMed] [Google Scholar]
- 11.Pierson CA, Jia N, Mo C, Lees ND, Strum AM, Eckstein J, Barbuch R, Bard M. Isolation, characterization, and regulation of the Candida albicans ERG27 gene encoding the sterol 3-keto reductase. Med. Mycol. 2004;42:461–473. doi: 10.1080/1369378032000141471. [DOI] [PubMed] [Google Scholar]
- 12.Thoma R, Schulz-Gasch T, D'Arcy B, Benz J, Aebi J, Dehmlow H, Hennig M, Stihle M, Ruf A. Insight into steroid scaffold formation from the structure of human oxidosqualene cyclase. Nature. 2004;432:118–122. doi: 10.1038/nature02993. [DOI] [PubMed] [Google Scholar]
- 13.Huff MW, Telford DE. Lord of the rings—the mechanism for oxidosqualene:lanosterol cyclase becomes crystal clear. Trends Pharmacol. Sci. 2005;26:335–340. doi: 10.1016/j.tips.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 14.Teske B, Taramino S, Bhuiyan MSA, Kumaraswami NS, Randall SK, Barbuch R, Eckstein J, Balliano G, Bard M. Genetic analyses involving interactions between the ergosterol biosynthetic enzymes, lanosterol synthase (Erg7p) and 3-ketoreductase (Erg27p), in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta. 2008;1781:359–366. doi: 10.1016/j.bbalip.2008.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Milla P, Athenstaedt K, Viola F, Oliaro-Bosso S, Kohlwein SD, Daum G, Balliano G. Yeast oxidosqualene cyclase (Erg7p) is a major component of lipid particles. J. Biol. Chem. 2002;277:2406–2412. doi: 10.1074/jbc.M104195200. [DOI] [PubMed] [Google Scholar]
- 16.Taramino S, Valachovic M, Oliaro-Bosso S, Viola F, Teske B, Bard M, Balliano G. Interactions of oxidosqualene cyclase (Erg7p) with 3-keto reductase (Erg27p) and other enzymes of sterol biosynthesis in yeast. Biochim. Biophys. Acta. 2010;1801:156–162. doi: 10.1016/j.bbalip.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JD, Smith J, Struhl K. Current protocols in molecular biology. Wiley-Interscience; NY: 1999. [Google Scholar]
- 18.Chan BE, Knowles BR. A solvent system for delipidation of plasma or serum without protein precipitation. J. Lipid Res. 1976;17:176–181. [PubMed] [Google Scholar]
- 19.Balliano G, Viola F, Ceruti M, Cattel L. Inhibition of sterol biosynthesis in Saccharomyces cerevisiae by N,N-diethylazasqualene and derivatives. Biochim. Biophys. Acta. 1988;959:9–19. doi: 10.1016/0005-2760(88)90144-0. [DOI] [PubMed] [Google Scholar]
- 20.Oliaro-Bosso S, Viola F, Matsuda SPT, Cravotto G, Tagliapietra S, Balliano G. Umbelliferone aminoalkyl derivatives as inhibitors of oxidosqualene cyclases from Saccharomyces cerevisiae, Trypanosoma cruzi, and Pneumocystis carinii. Lipids. 2004;39:1007–1012. doi: 10.1007/s11745-004-1323-2. [DOI] [PubMed] [Google Scholar]
- 21.Peterson GL. A simplification of the protein assay method of Lowry et al. which is more generally applicable. Anal. Biochem. 1977;83:346–356. doi: 10.1016/0003-2697(77)90043-4. [DOI] [PubMed] [Google Scholar]
- 22.Gachotte D, Eckstein J, Barbuch R, Hughes T, Roberts C, Bard M. A novel gene conserved from yeast to humans is involved in sterol biosynthesis. J. Lipid Res. 2001;42:150–154. [PubMed] [Google Scholar]
- 23.Morand OH, Aebi JD, Dehmlow H, Ji YH, Gains N, Lengsfeld H, Himber J. Ro 48-8071, a new 2,3-oxidosqualene:lanosterol cyclase inhibitor lowering plasma cholesterol in hamsters, squirrel monkeys, and minipigs: comparison to simvastatin. J. Lipid Res. 1997;38:373–390. [PubMed] [Google Scholar]
- 24.Oliaro-Bosso S, Taramino S, Viola F, Tagliapietra S, Ermondi G, Cravotto G, Balliano G. Umbelliferone aminoalkyl derivatives as inhibitors of human oxidosqualene-lanosterol cyclase. J. Enz. Inhib. Med. Chem. 2009;24:589–598. doi: 10.1080/14756360802318688. [DOI] [PubMed] [Google Scholar]
- 25.Milla P, Viola F, Ceruti M, Rocco F, Cattel L, Balliano G. 19-Azasqualene-2,3-epoxide and its N-oxide: metabolic fate and inhibitory effect on sterol biosynthesis in Saccharomyces cerevisiae. Lipids. 1999;34:681–688. doi: 10.1007/s11745-999-0413-5. [DOI] [PubMed] [Google Scholar]
- 26.Hughes AL, Todd BL, Espenshade PJ. SREBP Pathway Responds to Sterols and Functions as an Oxygen Sensor in Fission Yeast. Cell. 2005;120:831–842. doi: 10.1016/j.cell.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 27.Osborne TF, Espenshade PJ. Evolutionary conservation and adaptation in the mechanism that regulates SREBP action: what a long, strange tRIP it's been. Genes Dev. 2009;23:2578–2591. doi: 10.1101/gad.1854309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Albertini C, Leroux P. A Botrytis cinerea putative 3-keto reductase gene (ERG27) that is homologous to the mammalian 17b-hydroxysteroid dehydrogenase type 7 gene (17b-HSD7) Eur. J. Plant Pathol. 2004;110:723–733. [Google Scholar]
- 29.Fillinger S, Leroux P, Auclair C, Barreau C, Al Hajj C, Debieu D. Genetic analysis of fenhexamid-resistant field isolates of the phytopathogenic fungus Botrytis cinerea. Antimicrob. Agents Chemother. 2008;52:3933–3940. doi: 10.1128/AAC.00615-08. [DOI] [PMC free article] [PubMed] [Google Scholar]