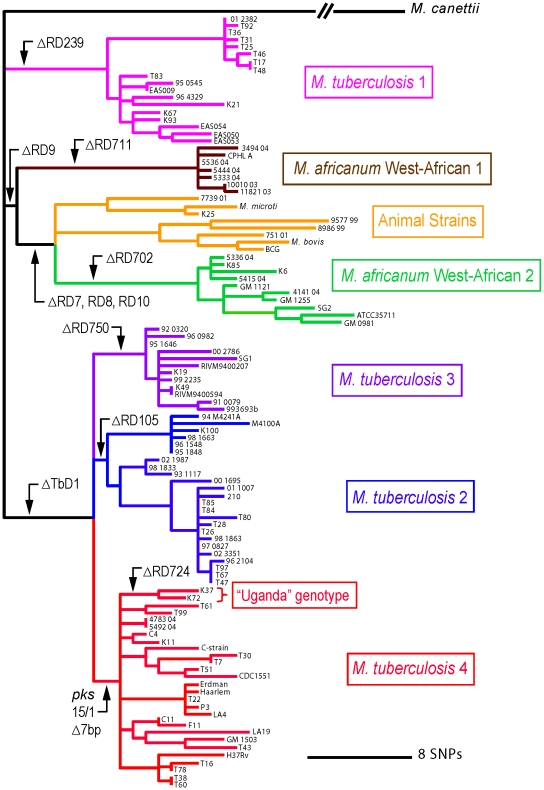
Figure 2. The position of M. africanum in the global phylogeny of the M. tuberculosis complex (MTBC) as originally published by Hershberg et al.[23].
This phylogeny is based on over 65 kb of DNA sequence data (89 concatenated gene sequences) in each of 108 strains of the MTBC and was inferred using maximum parsimony, which resulted in a single tree with negligable homoplasy [23]. Analysis by the neighbor-joining method resulted in an identical tree topology with high statistical support for all main branches [23]. This phylogeny has been referred to as the most robust and most detailed phylogeny of the MTBC to date, and thus should be considered as the new gold standard for classification of the MTBC [87]. The six main MTBC lineages adapted to humans and the animal strains are indicated in different colors. The human MTBC lineages include four M. tuberculosis lineages and the two M. africanum type I lineages. The “Uganda” genotype (formally referred to as M. africanum type II), which is a sub-lineage within M. tuberculosis lineage 4 (also known as the Euro-American lineage), is also shown. These lineages are completely congruent to previous classifications based on LSPs [15], [23], [78]. Black arrows indicate genomic regions or regions of difference (RDs) that are deleted in all descendent strains belonging to a particular lineage or sub-lineage. The scale indicates the genetic distance as number of SNPs (adapted from Figure 1 in [23] with additional data from [5]).