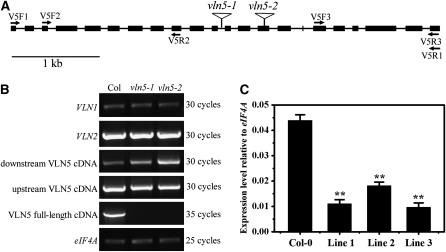
Figure 2.
Identification of Transcript Levels in T-DNA Insertion Mutants and Transgenic RNAi Lines.
(A) Physical structure of the Arabidopsis VLN5 gene. VLN5 contains 22 exons and 21 introns, which are represented by filled boxes and lines, respectively. The position of two independent T-DNA insertion mutants, designated vln5-1 (SAIL_512_F03) and vln5-2 (GABI_225F09), are noted by triangles above the diagram.
(B) Three separate pairs of primers, marked by arrows in (A), were designed to identify the level of VLN5 transcripts. The first pair of primers (V5F1 and V5R1) was designed to amplify VLN5 full-length cDNA, which was not present in vln5-1 and vln5-2 mutant plants. The second and third primer pairs, V5F2/V5R2 and V5F3/V5R3, were used to amplify upstream and downstream transcripts, respectively. Flowers from wild-type Col-0 plants and the two homozygous insertion lines were subjected to RT treatment. eIF4A was used as an internal loading control.
(C) VLN5 transcripts were reduced significantly in VLN5 RNAi flowers. Flowers from wild-type Col-0 and VLN5 RNAi plants were subjected to quantitative real-time PCR analysis. The expression level of the eIF4A gene was used as an internal control. All data represent the mean value of three biological replications. Error bars represent ± sd (n = 3); **P < 0.01.