Figure 4.
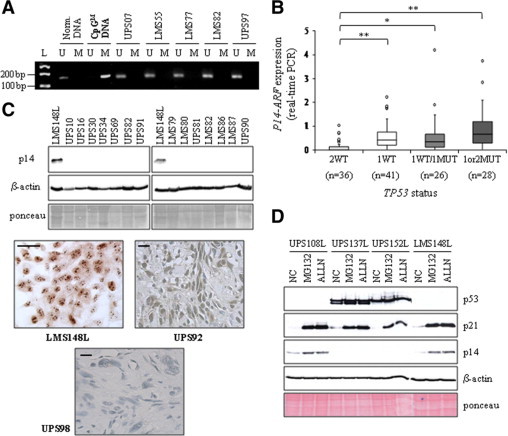
p14 expression in sarcomas. A: Some tumor methylation profiles of the p14 promoter are presented as examples. M: methylated-specific PCR, U: unmethylated-specific PCR. Normal DNA and CpGenome Universal Methylated DNA (CpGM DNA) are used as controls. L, molecular weight ladder. B: Boxplot analysis of p14 expression (real-time PCR). *P < 0.05; **P < 0.01 (Student's t-test) (2WT vs. 1WT: P = 5.7.10−5; 2WT vs. 1WT/1MUT: P = 2.10−2; 2WT vs. 1or2MUT: P = 1.5.10−4). Data for the two tumor types have been grouped because of the similarity of their results. C: Representative p14 Western blot. β-actin and Ponceau staining are shown as loading controls. Examples of p14 immunohisto/cytolabeling are presented. Scale bar = 40 μm. D: p14 Western blot performed after 8 hours of MG132 or ALLN treatment of four sarcoma cell lines. NC: negative control, cell lines cultured in medium with dimethyl sulfoxide. Features of these cell lines are similar to those of the corresponding tumors and are described in the Supplemental Table S4, at http://ajp.amjpathol.org. p21 and p53 are used to see the efficiency of the inhibitors. Ponceau staining and β-actin, loading controls.
