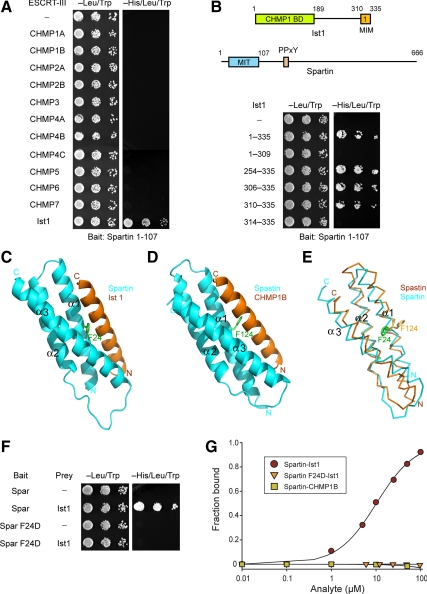
Figure 2.
Spartin MIT domain binds selectively to Ist1. (A) Yeast two-hybrid interactions between the spartin MIT domain bait and the indicated ESCRT-III prey constructs were assayed using the HIS3 reporter (sequential 10-fold yeast dilutions are shown). (B) Spartin MIT bait was tested for yeast two-hybrid interactions as described in A with the indicated Ist1 prey constructs. Boundary amino acid residues are indicated. Schematic diagrams for Ist1 and spartin are shown at the top, with amino acid residues numbered. BD, binding domain. (C) Structural model of spartin MIT domain with Ist1 MIM. Spartin is colored in cyan and Ist1 in orange. Residue Phe24 (F24) in the spartin MIT domain is shown in green. (D) Crystal structure of spastin MIT in complex with CHMP1B, from Yang et al. (2008). Spastin is colored in cyan and CHMP1B in orange. Residue Phe124 (F124) in the spastin MIT domain is colored in green. (E) Structural alignment model for the spartin (cyan) and spastin (orange) MIT domains. Spartin residue F24 is colored in green and spastin residue F124 is in yellow. (F) Wild-type and mutant spartin F24D MIT baits were tested for yeast two-hybrid interactions with Ist1. BD, binding domain. (G) SPR analysis of wild-type or mutant spartin F24D MIT domain (analyte) binding to immobilized Ist1-CTR or CHMP1B-CTR.