Fig. 3.
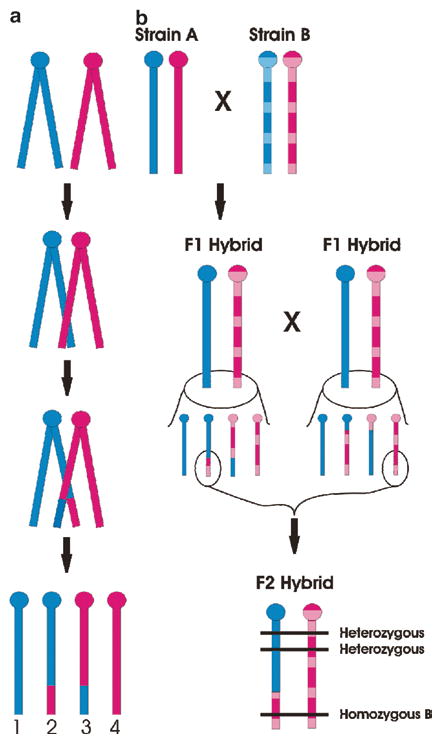
Linkage mapping theory. Note that murine chromosomes are telocentric, with the centromere (ball) located at the proximal end of the chromosome. Blue and pink colors represent the paternally and maternally derived chromosomes, respectively. For simplicity only one chromosome is illustrated. Graph A shows homologous recombination (crossing-over) in a chromosome during meiosis. During prophase I, the two paired chromosomes (i.e., four chromatids) are found very close to one another, and homologous sites on two chromatids can mesh and exchange places. As a result, four gametes are possible: two nonrecombinant (gametes 1 and 4), inherited unchanged from the parental form, and two recombinant (gametes 2 and 3), in which both paternal and maternal genetic information is represented. Graph B illustrates how homologous recombination produces “linkage” of intra-chromosomal genetic locations in an F2 hybrid mouse. Inbred Strain A (solid) and Strain B (hatched) when mated will produce identically heterozygous F1 hybrid animals. In the F1 hybrids, however, the effects of homologous recombination can be seen, and both nonrecombinant and recombinant gametes are created. Assuming the highlighted gametes result in the conception of an F2 hybrid, shown, genomic locations (horizontal lines) near each other physically will tend to have the same genotype (in this case, heterozygous), whereas genomic locations far away physically will tend to have a different genotype (in this case, homozygous for Strain B alleles). Thus, physical locations of unknown genetic factors can be estimated (see text)
