Abstract
The molecular mechanism(s) underlying transition from CCR5 to CXCR4 usage of subtype C viruses remain largely unknown. We previously identified a subtype C HIV-1 infected child whose virus demonstrated CXCR4 usage along with CCR5 upon longitudinal follow-up. Here we delineated the molecular determinants of Env involved in expanded coreceptor usage. Residue changes in three positions of Env V3 domain are critical for the dual-tropic phenotype. These include: substitution of arginine at position 11, MG or LG insertion between positions 13 and 14, and substitution of threonine at the position immediately downstream of the GPGQ crown. Introducing these mutations into V3 region of another R5 virus also conferred dual tropism. Molecular modeling of V3 revealed a possible structural basis for the dual-tropic phenotype. Determining what defines a subtype C X4 virus will lead to a better understanding of subtype C HIV-1 pathogenesis, and will provide important information relevant to anti-retroviral therapy.
Keywords: HIV-1 subtype C, envelope glycoproteins, Coreceptor switch
Background
HIV-1 enters host cells through a complex and dynamic process whereby envelope (Env) glycoprotein 120 (gp120) binds sequentially to CD4 and a coreceptor, typically CCR5 or CXCR4. Based on coreceptor usage in vitro, the majority of HIV-1 variants can be classified into three categories: CCR5-using viruses were designated as R5, CXCR4-using viruses, X4, and viruses able to use both receptors, R5X4. These patterns of coreceptor usage usually correspond to the phenotypes defined by the formation of syncytia in MT-2 cell line, where syncytia-inducing (SI) viruses were able to use CXCR4, while nonsyncytia-inducing (NSI) viruses used CCR5. Transmission of HIV-1 is, in general, associated with viruses that utilize CCR5, which predominate during the acute and asymptomatic phases of infection. Disease progression is often related to the emergence of viruses that have acquired the ability to use CXCR4. The appearance of CXCR4-using viruses was associated with an accelerated rate of loss of CD4+ T cells and a relatively rapid progression to AIDS and death.
Coreceptor usage and switching have been extensively studied in subtype B HIV-1 isolates. Some of the features of the Env glycoprotein involved in coreceptor usage have been revealed. It is known that HIV-1 cellular tropism and coreceptor specificity are largely determined by the third hypervariable loop (V3) of the viral gp120 glycoprotein, and distinct changes in this region and higher positive charges are associated with CXCR4 coreceptor usage among subtype B viruses. In addition to V3, other regions in gp120, such as V1/V2 and C4, have also been implicated in CXCR4 usage. A recent study indicated that gp41 transmembrane subunit was also involved in CXCR4 utilization.
Subtype C HIV-1 is currently responsible for the vast majority of new HIV-1 infections worldwide and is now the most prevalent subtype (http://www.unaids.org). In sub-Saharan Africa, subtype C HIV-1 accounts for more than 60% of infections and a significant number of infections are in infant and children. However, factors that contribute to the continuing expansion of subtype C viruses have yet to be clearly elucidated. Prior studies have suggested that CXCR4-using viruses were infrequently identified in subtype C HIV-1 infection compared to subtype B. Consequently, molecular mechanism(s) underlying the transition from R5 to X4 usage of subtype C virus remains largely unknown. However, with the evolving global HIV-1 epidemic, accumulating evidence has now shown that there is an increase in frequency in the detection of CXCR4-using subtype C HIV-1. A recent study from South Africa reported that CXCR4-using viruses were detected in 30% of a drug-naïve AIDS cohort, suggesting an ongoing dynamic evolution and maturing of the subtype C epidemic in South Africa. In addition, the emergence of the X4 viruses was found in about 50% subtype C infected patients receiving partially suppressive antiretroviral therapy (ART). This suggests that ART could promote the emergence of CXCR4-using virus. Although the sample size is relatively small in those studies, the introduction of antiretroviral therapy is likely to change the subtype C HIV-1 epidemic in Africa and the coreceptor usage of this subtype.
To date most of the studies regarding subtype C coreceptor switching mainly focus on genetic characterization of the viruses, and only a few documented the biological properties of these viruses. Moreover, the prediction of coreceptor tropism was mainly derived from the quasispecies rather than cloned viruses, and the molecular determinants that could govern subtype C HIV-1 coreceptor switch have not yet been deciphered experimentally. The appearance of X4 virus has been associated with poor clinical prognosis, and treatment with CCR5 inhibitors may select for CXCR4-using viruses, therefore, determining what defines a subtype C X4 virus and phenotypic characteristics of the virus will lead to a better understanding of the factors involved in pathogenesis and disease progression of subtype C HIV-1, and will provide important information relevant to anti-retroviral therapy as well.
We have previously characterized the longitudinal variations of the HIV-1 viral envelope glycoprotein obtained from a panel of HIV-1 infected children over a four-year follow-up period, and have identified a subtype C HIV-1 perinatally infected child whose virus had demonstrated CXCR4 usage in addition to CCR5 at 48 months after birth. This case provides us with an invaluable resource to study the evolution of coreceptor usage and to elucidate the molecular determinants of CXCR4 usage in the context of subtype C HIV-1 in an anti-retroviral naïve individual. To understand the basis for the expanded coreceptor usage, here we analyzed the sequence variations of the viral Env from the longitudinal follow-up samples and identified the molecular determinants of subtype C HIV-1 coreceptor transition from R5 to R5X4.
Materials and Methods
Cells
293 T, TZM-bl, MT-2, and Ghost 3 or U373-MAGI cell lines which express specific coreceptor were obtained from the NIH AIDS Research and Reference Reagent Program. 293 T and TZM-bl cells were maintained in Dulbecco’s modified Eagle medium (DMEM) with 10% fetal bovine serum (FBS), 100 U/ml penicillin and 100 μg/ml streptomycin. MT-2 cells were maintained in RPMI 1640 medium with 10% FBS, 100 U/ml penicillin and 100 μg/ml streptomycin. Ghost 3 cells expressing CD4 and either CCR5 or CXCR4 were maintained in DMEM with 10% FBS, 500 μg/ml G418, 100 μg/ml hygromycin B, penicillin-streptomycin and 1.0 μg/ml puromycin. U373-MAGI-CCR5 and U373-MAGI-CXCR4 cells were propagated in selection medium consisting of DMEM supplemented with 10% FBS, 200 μg/ml G418, 100 μg/ml hygromycin B and 1.0 μg/ml puromycin. Culture medium for virus infection and inhibition assays consisted of DMEM with 10% FBS.
Env cloning, plasmid construction, and mutagenesis
The existing, genetically characterized Env V1–V5 clones from child 1690 were the source of genetic materials for generation of the Env chimeras. Multiple representatives of Env clones with the signature sequences were selected from time points before (42 months) and after (48 months) coreceptor switch. The chimeric Env expression construct or proviral expression construct containing patient-derived Env V1–V5 region was generated as described previously. Briefly, Env V1–V5 fragment from those selected clones was first introduced into an Env expression vector, pSRH NLA/S/Av, with the subtype B (NL4-3) backbone. All the patient-derived chimeric Env expression constructs were first screened for biological function using the fusion assay as described previously. Finally, the functional envelope construct was transferred into a proviral expression vector-pNL4-3 A/S/Av, with NL4-3 viral backbone, resulting in the infectious molecular clones.
Single and combinations of mutations within V3 region of the virus were introduced by site-directed mutagenesis using QuickChange II XL kit (Strategene). All procedures were performed according to the manufacturer’s protocol. The V3 or V4 region chimerias were generated by overlapping PCR. Finally, the pSRH NLA/S/Av envelope construct of a serial of mutants were transferred into the proviral expression vector-pNL4-3 A/S/Av to generate the infectious molecular clones. The sequences of the resulting chimeras were confirmed by sequence analysis to ensure that only the desired substitutions were present.
Infectious HIV-1 viral stocks
Wild-type or mutated infectious molecularly cloned viral stocks were produced by transfection of 293 T cells with relevant proviral constructs. Nine μg of proviral constructs were transfected into 2.2 × 106 293 T cells using Fugene 6 (Roche). The resulting infectious viruses were harvested 48 h post-transfection. Tissue culture dose for 50% infectivity (TCID50) was determined for each virus in TZM-bl cells. Viral titer was defined as TCID50/ml.
Determination of viral coreceptor utilization
The coreceptor utilization of these mutated chimeric viruses was first tested in Ghost 3 cells that express CD4 and specific coreceptor, either CCR5 or CXCR4. These cells contain HIV-2 LTR promoter cassettes that express green fluorescent protein (GFP) in response to stimulation with HIV-1 Tat. Equivalent infectious units, 400 TCID50, of each chimeric provirus from transfected 293 T cells as described above was added to triplicate wells in a 24-well plate containing 4×104 Ghost cells per well. Ghost cells were assayed for infection by the observation of GFP expression at 4–5 days post-infection. The CXCR4 utilization was confirmed by the appearance of syncytia in MT-2 cells. For MT-2 infection, 500 TCID50 of each chimeric provirus was added to triplicate wells in a 24-well plate containing 5×104 MT-2 cells per well. On day 6–7 post-infection, cells were scored for syncytia formation.
Entry inhibition assays
Entry inhibitors TAK 779 or JM-2987 (hydrobromide salt of AMD 3100) were obtained from the NIH AIDS research and Reference Reagent Program. Entry inhibition was examined in TZM-bl or U373-MAGI-CCR5 or CXCR4 indicator cells in 96-well plates. Briefly, the cells were pre-treated with 50 μl of 2 μM of TAK 779 or AMD 3100 for 30 min at 37°C before infection. One hundred fifty TCID50 of each virus (in 50 μl) was then added to TZM-bl cells in triplicate for 72 h at 37°C or 100 TCID50 of each virus was added to U373-MAGI-CCR5 or CXCR4 cells in the presence of 10 μg/ml of DEAE-dextran for 48 h at 37°C. At the end of the culture period, the cells were lysed and processed for luciferase activity (Luciferase Assay System, Promega, Madison, WI) in TZM-bl cells or β–galactosidase activity (Galaco-Star System, Applied Biosystems, Bedford, MA) in U373-MAGI-CCR5 or CXCR4 cells. The percentage of entry inhibition was calculated by the luciferase or β–galactosidase activity in the presence of the entry inhibitor relative to that in the absence of inhibitor. Background luminescence in uninfected wells was subtracted from all experimental wells.
Infection assays
One hundred fifty TCID50 of each virus was used to infect 1×104 U373-MAGI-CCR5 or CXCR4 cells in triplicate in the presence of 10 μg/ml of DEAE-dextran in a 96-well plate. At 48 hrs post-infection, the infection was quantified by measuring β–galactosidase activity as described above.
Positive selection analysis
To identify selection on individual codons within V3, we computed the mean ratio of nonsynonymous to synonymous nucleotide substitutions (dN/dS) per site using the single-likelihood ancestor counting (SLAC) method available in the Datamonkey web interface of the HY-PHY package. In each case, this analysis utilized the GTR model of nucleotide substitution and an input neighbor-joining tree.
Structure modeling of the V3 loop
Structure modeling of the V3 loop for the R5 variant 48m-30 and the dual tropic variant 48m-18 was performed using the SWISS-MODEL server [http://swissmodel.expasy.org/; ]. The server chose the crystal structure of HIV-1 JR-FL gp120 core protein as a structural template [Protein Data Bank ID 2B4C; G chain; ] for both variants. The root-mean-square deviation (RMSD) between the model and the template was 0.05 Å for both coordinates.
The electrostatic potential of variants 48m-30 and 48m-18 was calculated using PBEQ Solver on CHARMM-GUI [http://www.charmm-gui.org/;] by solving the Poisson-Boltzmann equation. Model visualizations were made using PyMOL.
Results
Variants with distinct changes in the Env V1–V5 regions appeared at the time of the detection of the virus with the expanded coreceptor usage
Child 1690 was found to be HIV-1 positive at 12 months after birth and anti-retroviral naïve throughout the study. The R5 coreceptor tropism was maintained at 12, 24, 36 and until 42 months after birth; after which, the viral isolate from 48 months was found to use CXCR4 in addition to CCR5, and infected both macrophages and MT-2 T lymphoblasts where it formed syncytia [ and data not shown]. We further tested the coreceptor preference of the primary isolates from 42 and 48-month in U373-MAGI indicator cells which express CD4 and either CCR5 or CXCR4. The 48-month isolate was able to infect either CCR5 or CXCR4-expressing cells, whereas the 42-month isolate only exhibited CCR5-mediated infection (Fig. 1). In order to identify the molecular determinants of the expanded coreceptor usage of the 48-month isolate, we analyzed changes of the viral Env sequences from 12, 24, 36, 42 and 48-month of follow-up. Sequence analysis of the Env V1-V5 region indicated that there were distinct changes in the Env glycoprotein, mainly in the V3 and V4 regions, among clonal variants before (12, 24, 36 and 42 months) and after (48 months) NSI to SI or CCR5 to CCR5/CXCR4 phenotype switch (Fig. 2 and data not shown).
Figure 1. Coreceptor usage of HIV-1 quasispecies from 42 and 48-month after birth of child 1690.
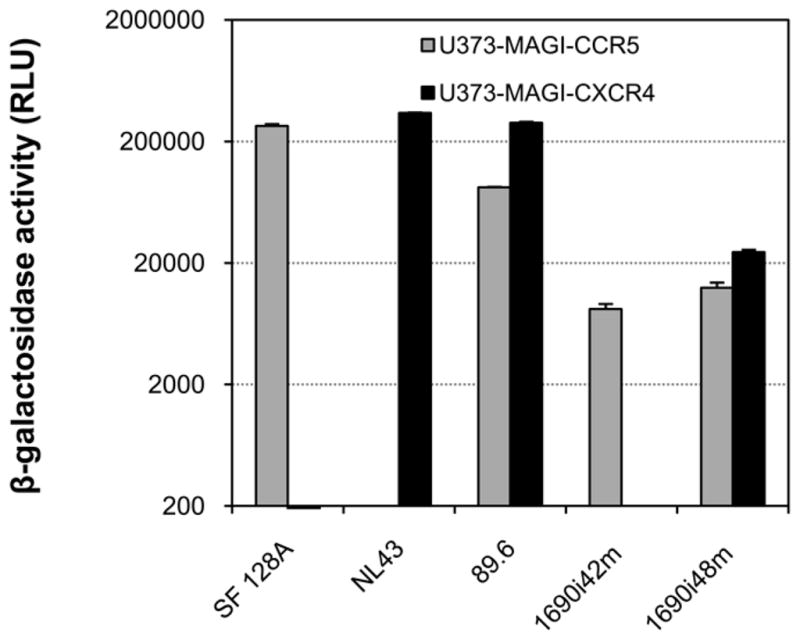
The infectivity mediated by the 42 or 48-month viral isolates was measured in U373- MAGI target cells expressing CD4 and either CCR5 or CXCR4. Infection with virus SF128A, NL4-3 or 89.6 served as controls for CCR5, CXCR4 or CCR5/CXCR4 usage virus, respectively. The results are representative of two independent experiments, and the error bars represent standard deviations.
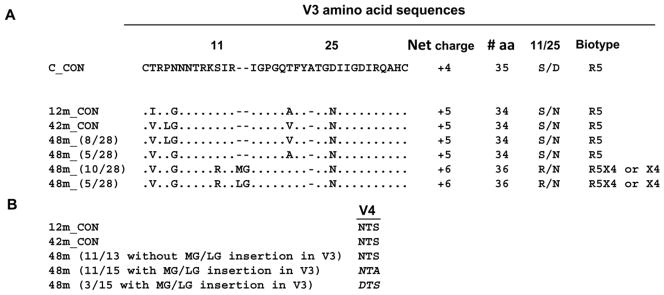
Figure 2. Comparison of sequences from child 1690 at different time points.
A. Consensus V3 amino acid sequence for variants at 12 and 42 months (12m_CON and 42m_ CON), respectively. Predicted V3 sequence for variants at 48 months (48m) with the number in parenthesis indicating the number of clones that contain the specific signature sequence. In all cases the sequences are compared to a HIV-1 subtype C consensus sequence. B. Comparison of the potential N-linked glycosylation site at the N-terminal of the V4 region. The loss of this glycan among all but one (14 out of 15) of the 48-month variants which have predicted CXCR4 usage based on V3 sequence analysis is in italics.
We first examined the sequence of the V3 region as this region significantly influences and determines coreceptor preference of non-subtype C HIV-1. Compared to the subtype C consensus, the V3 loops of all R5 viruses from 12 and 42-month were 34 amino acids in length and contained the GPGQ motif at the crown, characteristic of HIV-1 subtype C viruses (Fig. 2A). A neutral serine (S) residue was completely conserved at position 11 among 12 or 42-month variants. Sequence analysis of 48-month variants indicated that 46% sequences (13 of 28 clones sequenced) harbored the V3 region sequences which are similar to those of 12 or 42-month variants, with a net charge of +5 in the V3 region, indicative of R5 usage (Fig. 2A). However, the remaining 54% sequences (15 of 28 clones) of 48-month variants showed distinct changes in the V3 loop. Notably, an insertion of two amino acids –either MG or LG upstream of the GPGQ crown (between positions 13 and 14) was observed. In addition, mutations within the V3 region of these 48-month variants also include the change at position 11 from a neutral serine to a positive arginine (R), which was reported to associate with CXCR4 usage of subtype B HIV-1, and resulted in an increased positive charge (+6). Moreover, we also identified changes immediately downstream of the GPGQ crown from a valine (V) or alanine (A) to threonine (T). Interestingly, all the changes in the V3 domain as described above were linked to each other in 54% of sequences (15 of 28 clones) from 48-month variants, indicating possible functional association of these changes. Based on these observations, we anticipated that a mixture of R5 and either dual R5X4 or X4 variants could co-exist in the 48-month variants.
Moreover, we also determined whether non-V3 regions also possessed unique sequences suggestive of expanded coreceptor usage. Sequence analysis showed that there were some specific changes in the V4 region which co-evolved with the changes in the V3 region. In particular, a loss of a potential N-linked glycosylation site at the N-terminal of V4 region (mainly from NTS to NTA) was noted in all but one (14 out of 15 clones, 93%) variants which have predicted R5X4 or CXCR4 usage based on V3 sequence analysis (Fig. 2B). This N-linked glycan was completely conserved in 12-month sequences and maintained in the majority of the 42-month sequences and 48-month variants with predicted CCR5 utilization based on V3 sequence analysis (Fig. 2B). These findings on the association of the linked changes in the V3 and V4 regions suggest a possible functional relationship between the V3 and V4 regions.
Coreceptor usage of the molecularly cloned chimeric viruses
To test whether the characterized Env sequences possess the predicted coreceptor usage, we created chimeric proviral constructs by introducing subtype C Env V1–V5 fragments from child 1690 into a proviral expression vector with the subtype B (NL4-3) backbone. Multiple representative Env clones with the signature sequences as described above were selected from time points at 42- and 48-month before and after the expanded coreceptor usage.
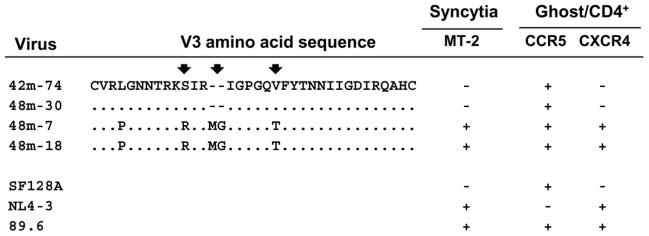
The coreceptor usage of the resulting representative chimeras was evaluated in indicator Ghost 3 cells. Infection with virus SF128A, NL4-3 or 89.6 served as controls for CCR5, CXCR4 or CCR5/CXCR4-using virus, respectively. The CXCR4 usage of the virus was confirmed by syncytium formation in MT-2 cells. Consistent with what we observed for the primary isolates, chimera 42m-74 which contains Env V1–V5 signature sequences from 42-month time point exhibited CCR5 tropism and did not form syncytia in MT-2 cells (Fig. 3). In contrast, chimeras derived from 48-month viral sequences bearing insertions in V3 and other linked changes in the V3 and V4 regions as described above (48m-7 and 48m-18) caused syncytia in MT-2 cells and could infect both CXCR4 and CCR5-expressing target cells (Fig. 3). However, Env chimeras which failed to infect MT-2 cells but could infect CCR5-expressing target cells were also identified among 48-month Env derivatives (Fig. 3, 48m-30). As expected, the CCR5-using 48-month chimeras share similar signature sequences with the 42-month chimeras (Fig. 3, 42m-74 vs 48m-30). Taken together, our phenotypic analyses of the 48-month sample revealed heterogeneous viral populations comprising of both R5 and R5X4 tropic variants.
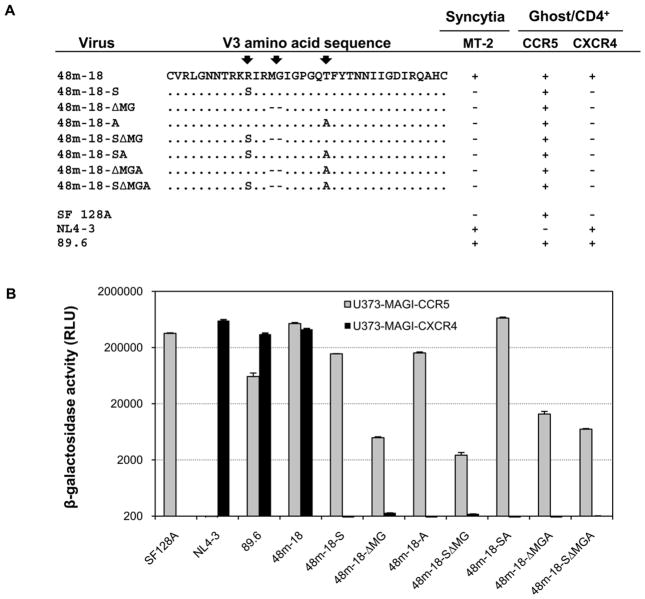
Figure 3. Amino acid comparison of the V3 sequences and coreceptor utilization of the representative viruses from 42 and 48-month.
Chimeric viruses and controls were produced by transfection of 293 T cells. An equal quantity of TCID50 for each virus was added to Ghost 3 cells expressing the indicated coreceptors. GFP expression induced by Tat activation of the integrated LTR-GFP cassette was used to assign positive or negative value after fluorescent microscopic examination at day 4 post-infection. MT-2 infection was monitored for syncytia formation at day 6 post-infection. Infection with virus SF128A, NL4-3 and 89.6 served as controls for CCR5, CXCR4 or CCR5/CXCR4 usage virus, respectively. Residue changes in the three positions that may affect the coreceptor usage are indicated by arrows.
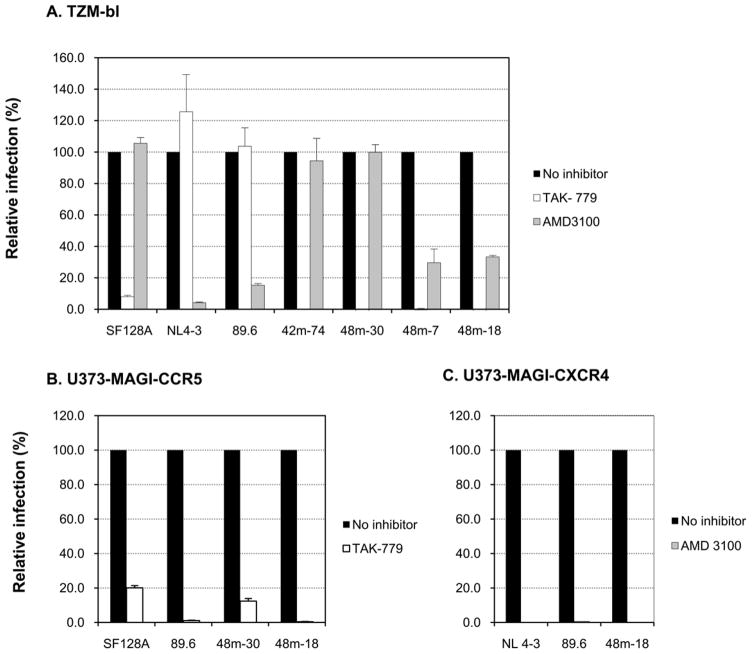
We further confirmed the coreceptor usage of the representative variants by infecting TZM-bl cells, which express both coreceptors, in the presence of a CXCR4 or CCR5 inhibitor (Fig. 4A). Infection of TZM-bl cells by control NL4-3 was efficiently blocked by the CXCR4 antagonist AMD3100 and unaffected by CCR5 inhibition, as expected for a CXCR4-dependent strain. In contrast, SF128 A was not affected by CXCR4 blockade but was blocked by the CCR5 antagonist TAK 779, as expected for an R5 strain. Consistent with observations by others, the dual tropic virus 89.6 was efficiently inhibited by AMD 3100 but not by TAK 779. Variants 18 and 7 from 48-month follow-up were almost completely blocked by the CCR5 inhibitor and were also inhibited by the CXCR4 blockade but less efficiently (70% inhibition), suggesting the dual CCR5/CXCR4 usage. In contrast, similar to 42-month variant 74, variant 30 from 48-month follow-up was essentially fully blocked by TAK 779 but not affected by AMD 3100, resembling the R5 phenotype (Fig. 4A). The dual tropic phenotype of variant 48m-18 was reinforced by the infection and blocking experiments in U373-MAGI indicator cells, where the virus was able to infect either CCR5 or CXCR4-expressing cells, and was completely inhibited by the cognate coreceptor inhibitor with the respective cell lines (Fig. 4B and C). Whereas, variant 48m-30 consistently showed CCR5-dependence as indicated by an effective inhibition of infection in U373-MAGI-CCR5 cells by TAK 779 (Fig. 4B).
Figure 4. Coreceptor preference of representative chimeric proviruses.
A. Coreceptor usage of 42-month and 48-month variants of child 1690 was determined by entry inhibition into TZM-bl cells with 1 μM CXCR4 inhibitor (AMD 3100) or CCR5 inhibitor (TAK-779). Viruses SF128A, NL4-3 and 89.6 served as controls for CCR5, CXCR4 or CCR5/CXCR4 usage virus, respectively. B, C. Infection and blocking with cognate coreceptor inhibitor were performed in U373-MAGI-CCR5 (B) or U373-MAGI-CXCR4 (C) cells. Viruses SF128A, NL4-3 and 89.6 served as controls for CCR5, CXCR4 or CCR5/CXCR4 usage virus, respectively. The data are expressed as mean values from experiments performed in triplicate, and the error bars indicate standard deviations.
V3 domain is necessary and sufficient to confer the expanded coreceptor usage of the 48-month isolate
Our sequence analyses have indicated that some potential sites in V3 and V4 regions may be involved in the expanded coreceptor usage of the 48-month isolate. To better understand the impact of the V3 and V4 regions on coreceptor switch, we constructed chimeric env clones by exchanging the V3 or V4 region between the 48-month dual CCR5/CXCR4-using variant 48m-18 and CCR5-using variant 48m-30 (Fig. 5A). The coreceptor usage of the resulting chimeras was evaluated by infection assays using Ghost 3 cells and MT-2 cells as described above. We found that the chimera 30-V3-18 which contains the V3 region from R5-tropic variant 48m-30 in the context of R5X4 env backbone (variant 48m-18) failed to infect CXCR4-expressing target cells, did not form syncytia in MT-2 cells, and only exhibited CCR5 tropism (data not shown). This suggests that the V3 domain of 48m-30 confers the CCR5 coreceptor usage.
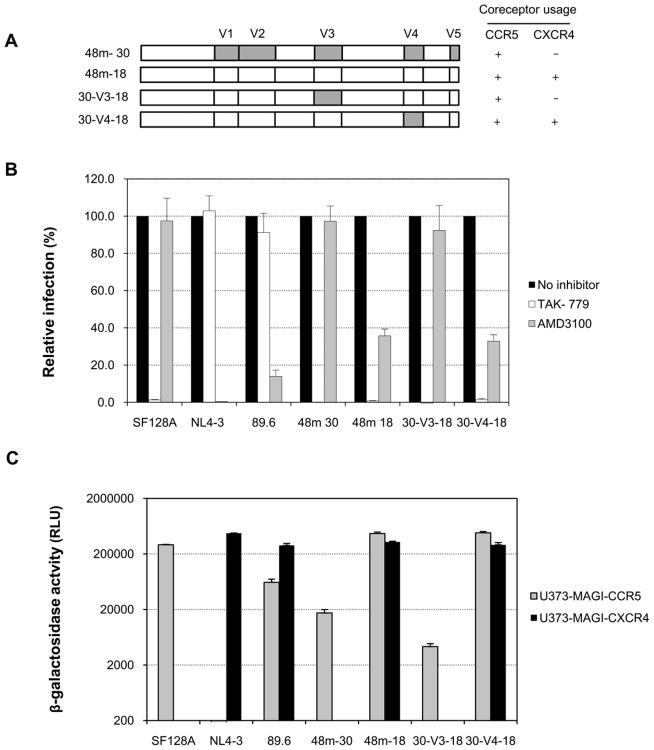
Figure 5. The impact of the V3 and V4 region on the coreceptor usage.
A. Schematic representation of the V3 or V4 domain-swapped chimeras. V3 or V4 region from R5 clone 48m-30 was introduced into the env backbone of R5X4 clone 48m-18 to generate the domain-swapped chimeras 30-V3-18 or 30-V4-18. The coreceptor utilization of each virus is indicated. B. Coreceptor usage of the 48-month parental variants (48m-30 and 48m-18) and env chimeras (30-V3-18 and 30-V4-18) was determined by entry inhibition into TZM-bl cells with 1 μM CXCR4 inhibitor (AMD 3100) or CCR5 inhibitor (TAK- 779). Viruses SF128A, NL4-3 and 89.6 served as controls for CCR5, CXCR4 or CCR5/CXCR4 usage virus, respectively. C. The infectivity mediated by the 48-month parental variants and the V3 or V4 domain-swapped chimeras was measured in U373- MAGI target cells expressing CD4 and either CCR5 or CXCR4. Infection with virus SF128A, NL4-3 or 89.6 served as controls for CCR5, CXCR4 or CCR5/CXCR4 usage virus, respectively. The results are representative of three independent experiments, and the error bars represent standard deviations.
We further tested the specificity of coreceptor usage of the chimera 30-V3-18 by infecting TZM-bl cells in conjunction with CXCR4 and CCR5 inhibitors. Infection with virus SF128A, NL4-3 or 89.6 served as controls for CCR5, CXCR4 or CCR5/CXCR4-using virus, respectively. Similar to the parental R5X4 variant 48m-18, the infection of TZM-bl cells by the chimera 30-V3-18 was essentially fully blocked by the CCR5 antagonist TAK 779 (Fig. 5B). However, the infection was not blocked by the CXCR4 antagonist AMD 3100, similar to that of the parental CCR5-using variant 48m-30 (Fig. 5B). We therefore concluded that V3 domain is necessary and sufficient to confer the expanded coreceptor usage of the 48m-18. In contrast, the chimera 30-V4-18, that contained the V4 region from R5-tropic variant 48m-30 in the context of R5X4 env backbone of variant 48m-18 remained R5X4 tropic, suggesting that V4 region is not essential for the expansion of coreceptor usage of the variant 48m-18 (Fig. 5B).
Infection of U373-MAGI target cells with the domain-exchanged chimeras provided additional evidence of the Env V3 domain as a major determinant of R5X4 usage. As shown in Fig. 5C, the chimera 30-V3-18 only displayed CCR5-mediated infection in CCR5-expressing target cells, albeit less efficient than the parental R5 tropic variant 48m-30. In contrast, the V4 domain-swapped chimera 30-V4-18 mediated similar level of infection in either CCR5 or CXCR4-expressing cells compared to the parental dual-tropic virus 48m-18 (Fig. 5C).
Changes in three amino acid residues of the Env V3 region are required for the expanded coreceptor usage
To identify the molecular determinants of CCR5/CXCR4 utilization, we performed site-directed mutagenesis to change the residues at the three positions (R, MG and T) in the V3 region of variant 48m-18 to resemble the amino acids observed in the R5 variants as shown in figure 6A. Since A and V co-existed among the R5 variants at similar frequency, we changed T to A for the convenience of mutagenesis. The coreceptor usage of each resulting mutant was first evaluated in Ghost 3 cells and MT-2 cells. Notably, when single or combinations of mutations in the V3 region were introduced, all the mutants failed to infect CXCR4-expressing target cells, and only exhibited CCR5 tropism. This is consistent with the observations that none of the mutants were able to form syncytia in MT-2 cells (Fig. 6A).
Figure 6. Single and combinations of mutations within the V3 region of R5X4 virus 48m-18 and coreceptor utilization of the resulting V3 mutant chimeric viruses.
A. GFP expression in Ghost 3 cells expressing the indicated coreceptors was used to assign positive or negative value after fluorescent microscopic examination at day 4 post-infection. MT-2 infection was monitored for syncytia formation at day 6 post-infection. Mutations in the three positions are indicated by arrows. B. Infectivity mediated by mutants containing single or combinations of mutations within the V3 region in the backbone of R5X4 virus 48m-18 was measured in U373- MAGI target cells expressing CD4 and either CCR5 or CXCR4. Infection with virus SF128A, NL4-3 or 89.6 served as controls for CCR5, CXCR4 or CCR5/CXCR4 usage virus, respectively. The results are representative of three independent experiments, and the error bars indicate standard deviations.
We further analyzed the infection of these mutants in U373-MAGI cells (Fig. 6B). As expected, the infection of all the mutants in 373-MAGI-CXCR4 cells was abrogated. Therefore, we conclude that, in the context of the 48m-18 V3 loop, the presence of R, MG and T was critical for envelope interaction with CXCR4 coreceptor, and the residues at those three positions are the minimal determinant for the CXCR4 usage.
Positively selected sites
To directly examine the molecular evolution of HIV-1 in the context of coreceptor usage, we estimated the selection pressure acting across residues within the V3 domain. Only two residues showed evidence for positive selection as indicated by the rate of nonsynonymous and synonymous substitutions (dN/dS). The residue immediately downstream of the GPGQ crown, which was an A, V or T was found to have a dN/dS ratio of 7.4 (p = 0.087). Furthermore, an S11R substitution had a dN/dS value of 4.50 albeit with a higher probability (p = 0.283).
V3 mutations in unrelated subtype C HIV-1 have similar effect on coreceptor usage
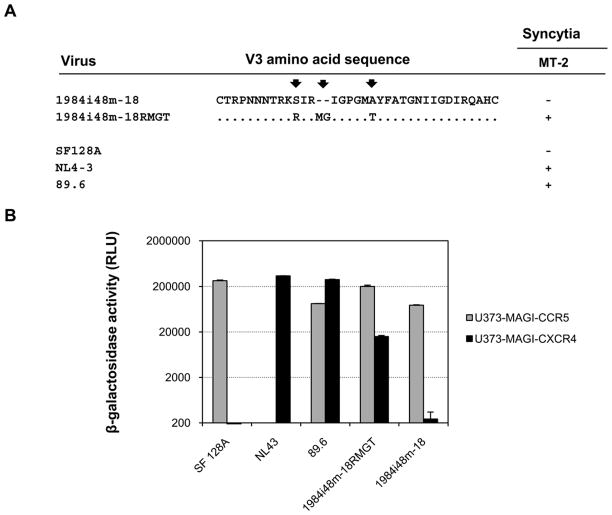
We next evaluated whether introduction of the mutations in the three positions in the V3 region could confer the dual tropic phenotype on a different HIV-1 genetic background. We selected another HIV-1 subtype C R5 variant, 1984i48m-18, that was derived from child 1984 at 48-month after birth, and introduced those mutations into its V3 region (Fig. 7A). We found that the mutations in the V3 region enabled the virus to form syncytia in MT-2 cells (Fig. 7A) and infect CCR5 or CXCR 4-expressing target cells (Fig. 7B). Thus, the presence of R, MG and T in the V3 region also conferred the dual tropism in a heterologous viral context.
Figure 7. Effects of V3 mutations on coreceptor usage of unrelated subtype C HIV-1.
A. The predicted V3 amino acid sequences of a 48-month variant of child 1984 and the V3 mutant. MT-2 infection was monitored for syncytia formation at day 6 post-infection. B. The coreceptor utilization of the 48-month parental variant and the V3 mutant was determined in U373-MAGI target cells expressing CD4 and either CCR5 or CXCR4. Infection with virus SF128A, NL4-3 and 89.6 served as controls for CCR5, CXCR4 or CCR5/CXCR4 usage virus, respectively. The results are representative of two independent experiments, and the error bars represent standard deviations.
V3 sequence comparison between the R5X4-using viruses in the present and published studies
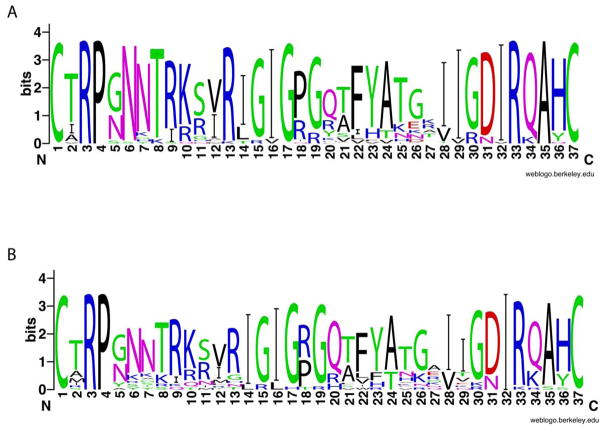
To address the biological relevance of our findings, and to further validate and confirm our results indicating that the V3 region is a major determinant for coreceptor usage of subtype C viruses, we compared the sequences of our R5X4 virus with those of subtype C R5X4 or X4 virus in the HIV-1 database or published literature to see whether the amino acids of interest are conserved. Sequence comparison revealed a strong homology in the V3 domain between our R5X4 virus and the published dual tropic or X4 tropic virus (Fig. 8). The two amino acid insertions upstream of the GPGQ crown (between positions 13 and 14) and the amino acid substitution of T immediately downstream of the GPGQ crown (position 21) were present in the majority of the sequences of R5X4 and X4 viruses. Nineteen of the 36 dual-tropic viruses (53%) had the two amino acid insertions, mainly IG, and 17 of the 36 viruses (47%) had the threonine substitution (Fig. 8A). A similar frequency in those changes was also observed for the X4 viruses, with 59% of X4 viruses (22 in total) harboring the two residue insertions and 50% of the viruses having the threonine substitution, respectively (Fig. 8B). The high sequence similarity in the V3 region among those CXCR4-using viruses indicated that the V3 region could be a major determinant that governs the coreceptor usage in a majority of the subtype C viruses with CXCR4 usage. Another important feature in the V3 region associated with CXCR4 usage is the arginine substitution in the GPGQ crown, mainly at position 18 or 20 (Fig. 8A and B) although this residue substitution is absent from our dual tropic viruses.
Figure 8. Sequence variation in the V3 region of HIV-1 subtype C R5X4 and X4 viruses from HIV-1 database and published literature.
The X axis indicates the amino acid position, whereas the symbol height (Y axis) indicates the relative frequency of each amino acid at that position of 36 R5X4 viruses (A) and 22 X4 viruses (B).
Molecular modeling of V3 structure
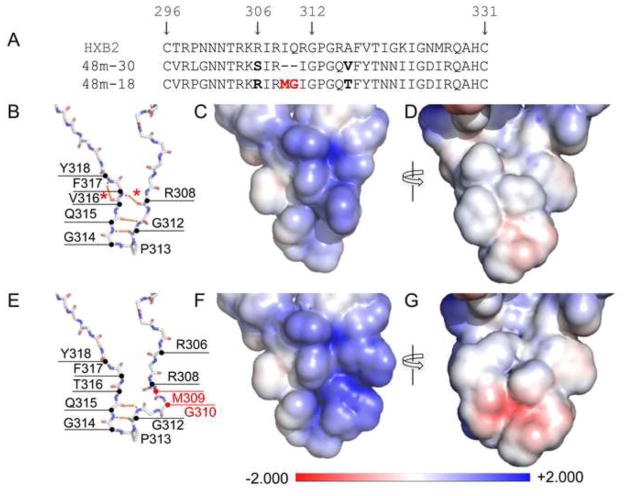
To evaluate the possible influences of those mutations observed in the V3 region on the structure of V3 in the context of gp120, we introduced the sequences of the 48-month dual tropic variant 48m-18 and CCR5-using variant 48m-30 into the X-ray crystal structures of V3-containing gp120 core, respectively (Fig. 9). Our structural model demonstrated that the MG insertion in variant 48m-18 could increase the bulkiness of the V3 tip and may introduce structural stress in the V3 tip. As a result, it could disrupt the hydrogen bond between F317 and R308, and the hydrogen bond between F317 and Y318, thereby changing the rigidity and increasing the flexibility of the V3 tip (Fig. 9E).
Figure 9. V3 sequence and structure modeling.
A. Alignment of V3 sequence of HXB2, 48m-30 and 48m-18 variant. The numbering of the gp120 residues corresponds to that of HXB2. The two residue insertions in 48m-18 are colored red. B–G. V3 structures of the R5 variant 48m-30 (B–D) and the dual tropic variant 48m-18 (E–G) are modeled with Swiss-Model (http://swissmodel.expasy.org) using 2B4C as the template. The protein backbone models show structural differences between variants 48m-30 (B) and 48m-18 (E). The insertions in 48m-18 are indicated by red annotation (M309 and G310). Hydrogen bonds within backbone atoms are indicated by the orange dashed lines. The asterisks indicate the hydrogen bonds lost in 48m-18 but present in 48m-30. The surface models show the electrostatic force distribution of variants 48m-30 (C, D) and 48m-18 (F, G). The models B and C or E and F are in the same view angles. Models D and G are rotated 180° about the vertical axis. The scale of the electrostatic forces is between-2.000 (red, negative charge) and +2.000 (blue, positive charge). The white area represents neutral charge.
We further compared the surface electrostatic potential of the two variants with different coreceptor preference (Fig. 9C–D, F–G). An extended positively charged area was observed in the V3 tip of the dual tropic variant 48m-18 due to the substitution of the positively charged arginine at position 306 (Fig. 9F versus 9C). In addition, a negatively charged cavity on the other side of the V3 tip, resulting from the MG insertion, was also apparent in the R5X4 variant 48m-18 compared to the R5 variant 48m-30 (Fig. 9G versus 9D). Therefore, the differences in the flexibility and charge distribution of the V3 tip may account for the observed coreceptor preference of these two variants.
Discussion
The pathogenic significance of coreceptor switch in HIV-1 infection is not completely understood. This situation is more complex in subtype C infection as limited information is available, especially regarding longitudinal isolates that showed coreceptor switching over time. In this study, we generated a set of molecularly cloned viruses from an infected child and delineated the molecular feature for the transition from CCR5 to dual CCR5/CXCR4 utilization of subtype C variants during longitudinal follow-up. However, we were not able to follow up the child beyond 48 months to determine whether the virus had eventually become exclusively X4 tropic.
The central role of the V3 loop in HIV-1 coreceptor usage has been well established for subtype B viruses. However, the genetic determinants associated with subtype C coreceptor utilization have not been defined at the clonal level, although several studies have described the potential molecular features, particularly in the V3 domain, that could influence subtype C virus coreceptor tropism. Our study, by swapping the V3 loop between the CCR5 and dual tropic variant, clearly indicated that consistent with non-subtype C viruses, V3 loop is a principal determinant for coreceptor usage of subtype C viruses. Therefore, the key role of V3 loop in governing HIV-1 coreceptor utilization could be a common mechanism among different subtypes, although regions other than V3 loop could also be involved in this process. In addition, we found that in the context of the dual tropic virus, V4 region is not essential for the expanded coreceptor usage. But it is possible that the co-evolving residues in V4 may act as compensatory or stabilizing mutations to accommodate some of the functional constraints imposed by the sequence variation observed in V3. Alternatively, the variations in V4 could directly or indirectly influence other biological properties of the Env. The significance of the co-variation of the V3 and V4 domains is currently under investigation.
We further analyzed the residues within V3 domain that were critical for the R5X4 dual tropic phenotype of the subtype C viruses. Different rules of estimating coreceptor tropism have been published based on the amino acid sequence of the Env V3 region. Among these, the most widely used is the 11/25 rule, where acquisition of positively charged residues at either amino acid position 11 or 25 in V3 was associated with CXCR4 usage. Recently, bioinformatics driven prediction systems have been developed. In this study, we found that a positively charged residue arginine was present at position 11 of the dual tropic variants. However, this positively charged residue is important (Fig. 6B, mutant 48m-18-S) but not sufficient (Fig. 6B, mutant 48m-18-ΔMGA) for CXCR4 utilization in the context of the variant 48m-18. In addition, the positively charged residue at position 25 was absent in all of the variants having the predicted dual tropic phenotype including 48m-18. Consistent with reports for subtype B, we observed a correlation between the increased positive charge of V3 loop and CXCR4 tropism, with the CXCR4-using viruses having higher positive charge (+6) compared to CCR5-using viruses (+5), suggesting a possible common feature for coreceptor switch among viruses from different subtypes. The second significant change in the V3 region contributed to CXCR4 usage was the insertion of either MG or LG between positions 13 and 14. Removal of this insertion abolished CXCR4 utilization by the virus (Fig. 6B), indicating the critical role of this insertion in the dual coreceptor tropism of subtype C viruses. One or two amino acid insertions at the same location have been observed in other studies on subtype C X4 viruses, but our study is the first to experimentally verify the significance of the insertions for the expanded CXCR4 tropism. Another genetic feature in the V3 region related to the dual tropic subtype C viruses was the amino acid substitution by T immediately downstream of the GPGQ crown. Similar to the other two mutations, this residue is required but insufficient in conferring the dual tropism of the viruses (Fig. 6B). The importance of this amino acid substitution in determining coreceptor preference has not been indicated in previous studies. However, A-to-T substitution has been implicated in escape from certain neutralizing antibody, and also found to contribute to the acquired replication in the thymus in vivo but not in macrophage. Interestingly, our selection analysis revealed that the amino acid substitution by T at this position was under strong positive selection, with an estimated dN/dS value of 7.4, suggesting that the presence of T at this site may be essential for the binding to both CCR5 and CXCR4. Therefore, the three mutations in the V3 region (R, MG and T) collectively contribute to CXCR4 engagement. More importantly, introducing the three mutations into an unrelated subtype C R5 variant was also sufficient in imparting the expanded CXCR4 tropism.
It has been suggested that coreceptor switching occurs less frequently for subtype C viruses compared to subtype B viruses. The intrinsic differences in V3 conformation between the two subtypes could be responsible for the limited CXCR4 adaptation for subtype C virus. It is also possible that the more complex genetic pathway with more than one mutation sites needed for efficient CXCR4 usage, as indicated in this study and those of others, may also contribute to the infrequent CXCR4 usage of subtype C viruses.
The structural basis for CCR5 or CXCR4 specificity is still poorly understood. The current model of how gp120 interact with CCR5 suggests that the tip of V3 region of gp120 binds to the second extracellular loop (ECL2) of CCR5, the base and stem of V3 and bridging sheet binds to the N-terminus of CCR5. The involvement of ECLs and the N-terminal of CXCR4 in the binding of CXCR4 was also observed. In this study, the mechanism by which the three mutations in the V3 loop determine CXCR4 usage is unclear. It is possible that the structural alterations created by MG insertion in the V3 tip may affect the interaction of the V3 tip with ECL2 of CCR5, and as a compensation of this, the insertion could potentially change the reliance of V3 base and bridging sheet of gp120 on the CCR5 N-terminus for entry. On the other hand, the changes in the structure that affect how V3 recognizes CCR5 may impart a more flexible conformation to the virus. This flexibility may be essential for engaging CXCR4 as an alternative coreceptor while retaining its ability to interact with CCR5. It has been suggested that dual-tropic HIV-1 strains may have extreme flexibility to display two different conformations or binding sites on the V3 surface in order to interact with either CCR5 or CXCR4. Our surface electrostatic potential analysis appeared to support this notion by showing the presence of both a negative cavity and a strong positively charged V3 tip in the dual tropic variant 48m-18. The positively charged surface may interact with the unusually anionic surface of CXCR4. On the other hand, the negative cavity in V3 seems to be consistent with the proposed negatively charged or neutral nature of R5 tropic viruses. Further dissecting the possible impact of the adapted changes associated with the expanded coreceptor utilization on the ability of V3 to interact with CCR5 or CXCR4 will provide important information on the molecular mechanisms and the structural basis for subtype C HIV-1 coreceptor usage.
In summary, our study provides strong evidence for the molecular determinants of subtype C HIV-1 dual coreceptor tropism. Specific changes in three areas in the V3 domain including residue substitutions or insertions account for the transition from a R5 to R5X4 phenotype. In addition, the increased net charge in V3 is another feature coupled with the CXCR4 utilization. Our structural modeling suggested possible structure alterations in V3 resulted from those sequence changes, and thus enabling the virus to adopt the structure required for the R5X4 phenotype. Although this is the only case of expanded coreceptor usage that was identified among our Zambian mother-infant cohort, to our knowledge our study is the first to use cloned viruses to experimentally decipher the molecular determinants of the expanded CXCR4 tropism of subtype C HIV-1 since previous studies are limited to sequence analyses only. In addition, our study suggested that the presence of the three mutations in the V3 region may also affect coreceptor usage of other subtype C viruses. Moreover, the strong homology in the V3 region, particularly the two amino acid insertions and the T substitution, between our R5X4 tropic virus and the published subtype C dual or X4 tropic virus sequences further strengthens our conclusions and reinforces the impact of these mutations on CXCR4 preference of subtype C viruses. With more subtype C X4 viruses being identified, particularly with the emerging availability of entry inhibitors, more detailed studies on virological adaptations within subtype C viruses associated with the acquired ability to use CXCR4 as a coreceptor will be needed to further elucidate the molecular mechanisms controlling coreceptor preference. The panel of genetically related but functionally distinct chimeric viruses should be useful tools in understanding how specific changes in the Env V3 domain favoring coreceptor switch may affect the biological properties of the virus, and contribute to the pathogenesis of subtype C HIV-1.
Acknowledgments
This study was supported by Public Health Service grants CA75903, Fogarty Training grant TW001429 and NCRR COBRE grant RR15635 to C.W.; Layman Award from the University of Nebraska-Lincoln and NCRR COBRE P20 RR15635 to H.Z.. We thank Lee Ratner (Washington University) for helpful discussions, Danielle Shea for assistance with flow cytometry and Sandra Gonzalez for assistance with part of sequencing.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.