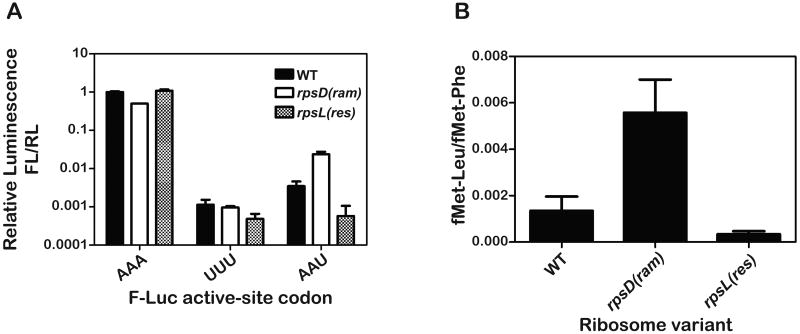
Figure 3. The rpsD(ram) and rpsL(res) variants exhibit the expected phenotype both in vivo and in vitro.
(A) The relative luminescence of firefly luciferase (F-Luc) active site (K529) variants to that of control renilla-luciferase (R-Luc) expressed in the indicated strains is plotted (normalized to K529 F-Luc value in the WT strain) (Kramer and Farabaugh, 2007). When the K529 codon (AAA) is replaced by the non-cognate phe codon (UUU), only background activity of F-Luc is observed in all strains. In contrast, when this codon is replaced by the near-cognate codon (AAU), activity associated with the error frequency of tRNALys reading the substituted AAU codon is observed.
(B) The error frequency of WT, rpsL(res) and rpsD(ram) ribosome variants in our in vitro system. The error frequency was determined on UUC programmed ribosomes with excess ternary complex containing equimolar concentrations of Phe-tRNAPhe and Leu-tRNA2Leu where the relative amounts of fMet-Leu and fMet-Phe dipeptide were determined. In both graphs, the mean of three independent experiments is plotted and the error bars represent standard deviations from the mean.