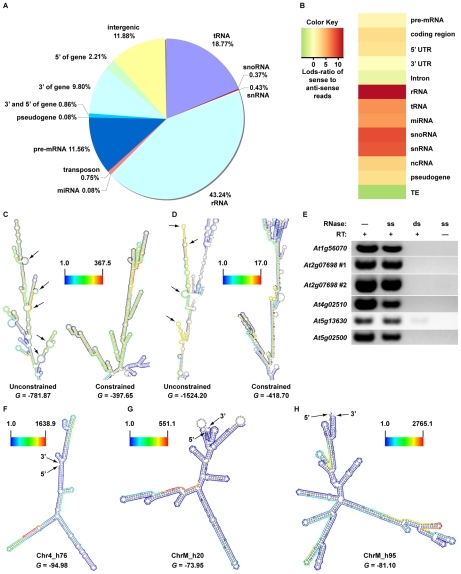
Figure 7. A sequencing-based approach to interrogate mRNA secondary structure genome-wide.
(A) Classification of genome-matching dsRNA-seq reads after two rounds of rRNA-depletions (2X Ribominus approach). (B) The heatmap indicates the strand bias of 2X Ribominus dsRNA-seq reads with respect to specific classes of RNA molecules. The color intensities indicate the degree of strand bias as specified by a normalized Lods-ratio value of sense/anti-sense mapping reads (red, sense; green, antisense; yellow, unbiased). TE, transposable element. (C, D) Models of secondary structure for Arabidopsis (C) At2g07698 and (D) At4g02510 transcripts determined by default (unconstrained) or dsRNA-seq constrained parameters for RNAfold (screenshots from the structural viewer at http://tesla.pcbi.upenn.edu/annoj_at9/). The sequences interrogated in (E) (At2g07698 #1 and At4g02510) are highlighted in yellow. The scale bar between the two models indicates the read counts that are normalized by the length of sequenced bases for the transcript. Black arrows indicate RNA loops that are >5 nt within the yellow shaded portions of the models. G denotes the Gibb's free energy value (kilocalories/mole) for the corresponding RNA secondary structure model. (E) Random-primed RT-PCR analysis of dsRNA ‘hotspots’ from At5g56070, At2g07698 (2), At4g02510, At5g13630, and At5g02500 after treatment of total RNA samples with either a single-stranded (ss) or double-strand RNase (ds). Samples that were not treated with reverse transcriptase (RT -) or either RNase (-) serve as controls for this experiment. (F–H) Models of secondary structure for Arabidopsis (D) chr4_h76 (chr4: nt 1476284–1476589), (E) chrM_h20 (chrM: nt 46875–47251), and (F) chrM_h95 (chrM: nt 334344–334833) novel intergenic transcripts determined by dsRNA-seq constrained parameters for RNAfold (screenshots from the structural viewer at http://tesla.pcbi.upenn.edu/annoj_at9/). The scale bar to the left (F, G) or right (H) of each model indicates the read counts that are normalized by the length of sequenced bases for the transcript. G denotes the Gibb's free energy value (kilocalories/mole) for the corresponding RNA secondary structure model.