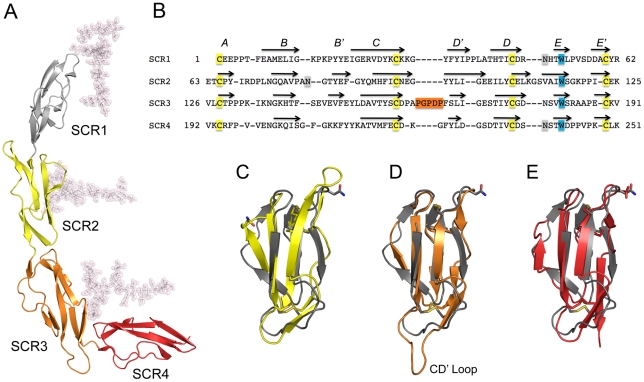
Figure 3. Structure of CD46-4D.
(A), Overall structure of CD46-4D, with domains SCR1-SCR4 shown in different colors. The protein carries glycosylation at positions Asn49 (SCR1), Asn80 (SCR2) and Asn239 (SCR4). Although only single NAG residues are visible at Asn49 and Asn80, more extensive glycosylation has been modeled to present a view of the protein that resembles its physiologic state (see Methods). (B) Structural alignment of all four repeats of CD46. The conserved cysteine and tryptophan residues, which are hallmarks of SCR domains, are highlighted in yellow and blue, respectively. The five-residue insertion of the unique CD' loop of SCR3 is shown in orange. Sites of N-linked glycosylation are highlighted in orange. Beta strands are indicated with arrows, and are labeled with letters. The alignment was performed with Modeller (http://salilab.org/modeller/modeller.html) using a gap penalty of 3. (C–E). Superpositions of domains SCR2 (yellow, panel C), SCR3 (orange, panel D) and SCR4 (red, panel E) onto SCR1 (grey). Side chains of conserved cysteine and tryptophan residues of each domain are shown in atomic detail to visualize the agreement of the core domains. Also shown as stick models are the three asparagine residues that carry glycosylation. The unique CD' loop in SCR3 is labeled.