Table 2. Regressions between life-history traits with and without correction for phylogenetic signal.
| Traits and dataset | Test | Slope (SE) | Y-intercept (SE) | ln(likelihood) | AIC |
| log(spVL) vs. dsCD4 (MSM strict) | OLS | −1.7e-3 (8.1e-4) (8.1e-4) |
4.26 (0.08) (0.08) |
−56 | 119 |
| RegBM | −2.7e-3 (6.8e-4) (6.8e-4) |
4.1 (0.34) (0.34) |
−57 | 121 | |
| Trait variation among risk groups (all liberal) | OLS | log(spVL) and dsCD4 and dsCD4
|
|||
| RegBM | log(spVL) and dsCD4 and dsCD4
|
The first line is a regression between log(spVL) and dsCD4. The second line tests if values for a given trait vary across risk groups. OLS is the ordinary generalised least square without phylogenetic correction (a generalised linear model yielded similar results); RegBM indicates a correction based on the tree assuming Brownian motion. SE stands for ‘Standard Error’. For further details, see the Supplementary Methods. Significance code for the p-value of the test is ‘ ’
’ 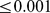 , ‘
, ‘ ’
’ 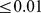 , ‘
, ‘ ’
’ 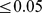 and ‘n.s.’ for non sognificant.
and ‘n.s.’ for non sognificant.
