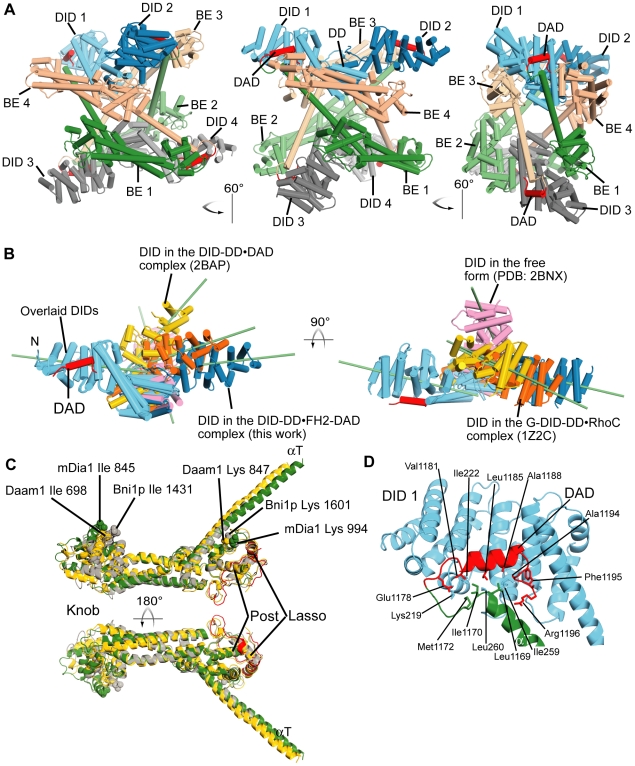
Figure 2. Crystal structure of autoinhibited tetrameric mDia1.
(A) Structure of tetrameric mDia1 in three orientations. Abbreviation: BE, bridge element. (B) Overlay of the N-terminal inhibitory domain (DID-DD) of mDia1 from various crystal structures: the DID-DD•FH2-DAD complex (blue, this work), the free DID-DD-CC (pink, PDB code:2BNX), the DID-DD•DAD complex (yellow, 2BAP), and the RhoC-G-DID-DD complex (orange, 1Z2C). One of the two DIDs in each dimeric DID-DD structure is superimposed (left, sky blue DID) to illustrate the differences in position of the other DIDs. For clarity, the structures solved as complexes with RhoC and DAD, are shown without these binding proteins. The long axis of each DID molecule is indicated as a green bar. (C) An overlay of the Cα models of the bridge elements of mDia1 (green for the core and red for the lasso regions, this work), Daam1 (yellow, PDB code: 2J1D) and Bni1p (gray, 1Y64) is shown. The key residues that are critical for the activities and are identified by earlier works on Bni1p yeast formin (Ile 1431 in the Knob site and Lys 1601 in the Post site) [20], [21], [41] are shown in sphere model with their side-chains. The lysines in the Post site are well superposed but isoleucines in the Knob site are not. (D) An expanded view of the αT-DAD binding to DID. The orientation of the figure is similar to the middle of (A). The dotted line indicates the hypothetical connecting linker (5 residues 1172-1176) whose electron density is missing. The Ile 1170 contacts the hydrophobic residues of DID and DAD so that the peak corresponding this residue in the NMR experiment shows a shift from the free to the complex form (see also Fig. 4).