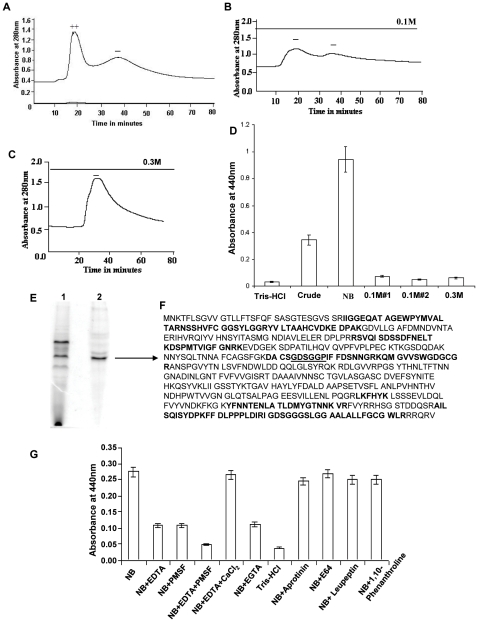
Figure 2. Partial purification and identification of protease.
Chromatographic profile of ammonium sulphate precipitated crude proteins from culture supernatants of CHA6.8ΔprtV strain loaded onto an anion exchange column (DE-52). A) Proteins eluted in the non-binding fraction (NB), B) proteins eluted with 0.1 M NaCl, C) proteins eluted with 0.3 M NaCl, +/− shows presence or absence of protease activity, D) azocasein assay with pooled samples (30 µg) NB, 0.1 M#1, 0.1 M #2, 0.3 M and crude proteins. E) Native PAGE profile (lane 1) of crude proteins of CHA6.8ΔprtV strain and (lane 2) of partially purified protease (NB) from DE-52 column. The marked protein band was analyzed by MS/MS sequencing and the peptides highlighted showed homology with a 59-kDa trypsin-like serine protease encoded by VC1649. F) The underlined GDSGGP are the amino acid sequences around the serine residue present in trypsin-like serine proteases. G) Protease inhibition test of NB fraction (5 µg) with protease inhibitors 10 mM EDTA, 25 mM PMSF, 25 mM PMSF and 10 mM EDTA, 10 mM EDTA and 20 mM CaCl2, 10 mM EGTA, 1 µg/ml aprotinin, 28 mM E64, 1 µg/ml leupeptin and 10 mM 1,10-phenanthroline incubated for 30 mins at 37°C. Residual protease activity was assayed by azocasein assay. Twenty-five mM Tris-HCl was used as a negative control. The values shown are the means with standard deviations from three experiments.