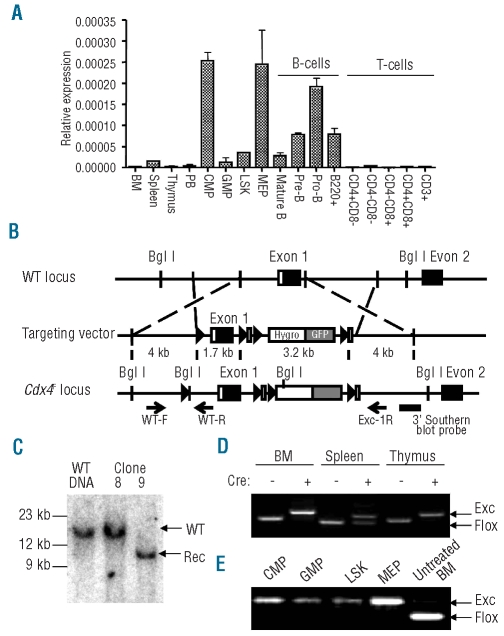
Figure 1.
Cdx4 expression and conditional inactivation strategy. (A) Quantitative real-time RT-PCR was used to measure Cdx4 mRNA expression levels relative to β-actin in normal hematopoietic tissues and flow sorted cells. BM: bone marrow; PB: peripheral blood; LSK, enriched for hematopoietic stem cells; CMP common myeloid progenitors; GMP, granulocyte-monocyte progenitors; MEP, megakaryocyte-erythroid progenitors. (B) Homologous recombination targeting strategy to obtain Cdx4F/F mice. (C) Southern blot analysis using the 3′ probe indicated in Panel B. WT: wild-type Cdx4 locus, Rec: Cdx4 locus target by the conditional knock-out construct. Clone 9 showed homologous recombination and was used to generate the conditional knock-out mouse line. Clone 8 was not correctly targeted, and demonstrates the germline configuration. (D) PCR to assess Cdx4 excision 5 weeks after pIpC treatment was performed on DNA from the indicated organs of Cdx4F/F Mx1Cre+ or Cdx4F/F Mx1Cre− animals using the three primers shown in Panel B (WT-F, WT-R, Exc-1R). Flox: unexcised allele, Exc: Excised allele. (E) LSK cells and myeloid progenitor populations from Cdx4F/F Mx1Cre+ were flow sorted 6 weeks after pIpC treatment and DNA was extracted to assess excision efficiency as described in (D). DNA from untreated Cdx4F/F Mx1Cre+ total bone marrow cells was used as a control.