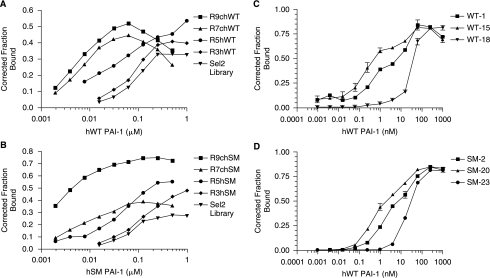
FIG. 1.
Systematic evolution of ligands by exponential enrichment (SELEX) yielded aptamers that bind to plasminogen activator inhibitor-1 (PAI-1) with high affinity. (A) The progress of the selection against the wild-type (WT) version of PAI-1 was followed using a nitrocellulose filter binding assay. Inverted triangles (▾) represent the starting RNA library (Sel2). Diamonds (♦) represent round 3, circles (•) represent round 5, triangles (▴) represent round 7, and squares (▪) represent round 9. The x-axis shows the human wild-type PAI-1 concentration and the y-axis shows the fraction of RNA bound to the protein. (B) The progress of the selection against the stable mutant (SM) version of PAI-1 was followed using a nitrocellulose filter binding assay. Inverted triangles (▾) represent the starting RNA library (Sel2). Diamonds (♦) represent round 3, circles (•) represent round 5, triangles (▴) represent round 7, and squares (▪) represent round 9. The x-axis shows the human stable mutant PAI-1 concentration and the y-axis shows the fraction of RNA bound to the protein. (C) Binding affinities of aptamers WT-1, WT-15, and WT-18 generated against WT PAI-1 were determined using a nitrocellulose filter binding assay. Squares (▪) represent WT-1, triangles (▴) represent WT-15, and inverted triangles (▾) represent WT-18. Each data point was performed in triplicate; error bars represent the standard error of the mean (SEM) of the data. (D) Binding affinities of aptamers SM-2, SM-20, and SM-23 generated against SM PAI-1 were determined using a nitrocellulose filter binding assay. Squares (▪) represent SM-2, triangles (▴) represent SM-20, and circles (•) represent SM-23. Each data point was performed in triplicate; error bars represent the SEM of the data.