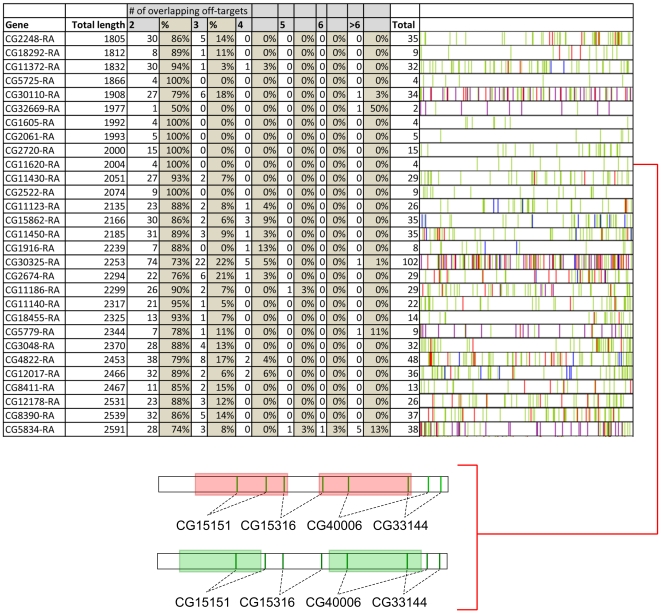
Figure 2. Non-overlapping cDNA sequences of randomly chosen genes have a high prevalence of sharing off-targets in pre-mature sequences of other genes.
Example of genes for which –based on sequence similarities- overlapping off-targets exist. The number of items per off-target group is given both in numbers and percentages. As an example: the cDNA of gene CG11372-RA contains 30 events of sequences with a shared off-target and 1 event of triplicate sequences that share the same off-target and 1 event of quadruple sequences that share the same off-target. Green lines represent sites that share an identical off-target with one other site, red lines represent sites that share an identical off-target with 2 other sites, and blue lines with 3 other sites. Purple lines represent sites that share identical off-targets with 5 or more other sites. The complete report from the 99 randomly selected genes is presented in Supporting Information S1. Note that for some genes the lines representing the off-target events are in close proximity and cannot be distinguished as separate lines in this illustrative figure. Overall, there appears to be a tendency for the occurrence of overlapping off-targets at the boundary (UTR's) of the genes, as is evident in for example CG5834 (last gene in the list). The insert shows a more detailed illustration for the analysis of the cDNA of gene CG11620. The green vertical lines represent sites that share an identical off-target with one other site. Sites that share the same off-target are connected with dotted lines and the shared off-target (as CG number) is indicated for each pair. To avoid off-target effects, dsRNA constructs should be chosen in such a way that the dsRNA constructs do not include both members of one pair. The green boxes represent areas which do not include both members of one pair. In order to reduce the likelihood of shared-off target effects, dsRNAs should be designed using sequences from the green regions. In contrast the red areas do include both members of one pair. When 2 independent dsRNA constructs will be designed from these areas, these dsRNA constructs do share sequence similarities with the same off-target gene. Our tool provides for all the genes present in the Drosophila genome the green areas.