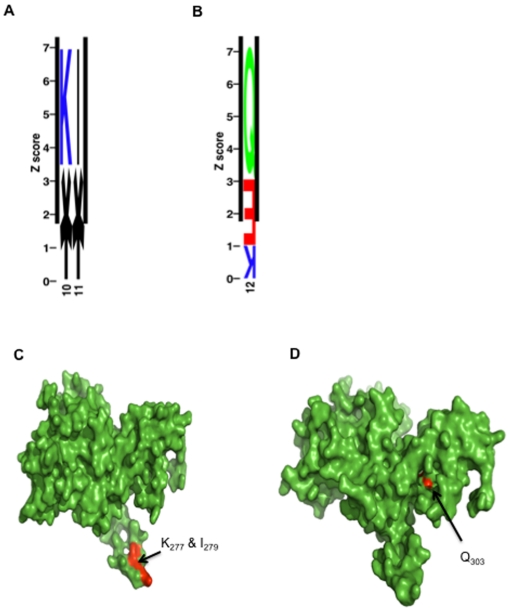
Figure 2. VAR2CSA DBL5ε patterns distribution.
(A, B): Sequence logo showing the identified significantly distributed residues I, K and Q The sequence logo displays the residues present at each position, where at least one residue was identified as being significantly distributed with respect to associated numerical parameter. Each letter denotes a given residue and the height corresponds to increasing z-score. The residues are coloured according to: Acidic [ED]: red, Basic [RKH]: blue, Neutral [GNQSTY] = green, Hydrophobic [ACFILMPVW] = black. Numbers below each column denotes corresponding position in the multiple alignment. Letters positioned correctly are associated with high values and upside down letters with low. An asterisk denotes a deletion. It should be noted that in the sequence logos other residues appears (*, E, K), these are however not identified as significantly distributed (i.e. p>0.05). DBL5ε models showing the position of the identified significant residues (red), T277, I279 (C) and Q303 (D).