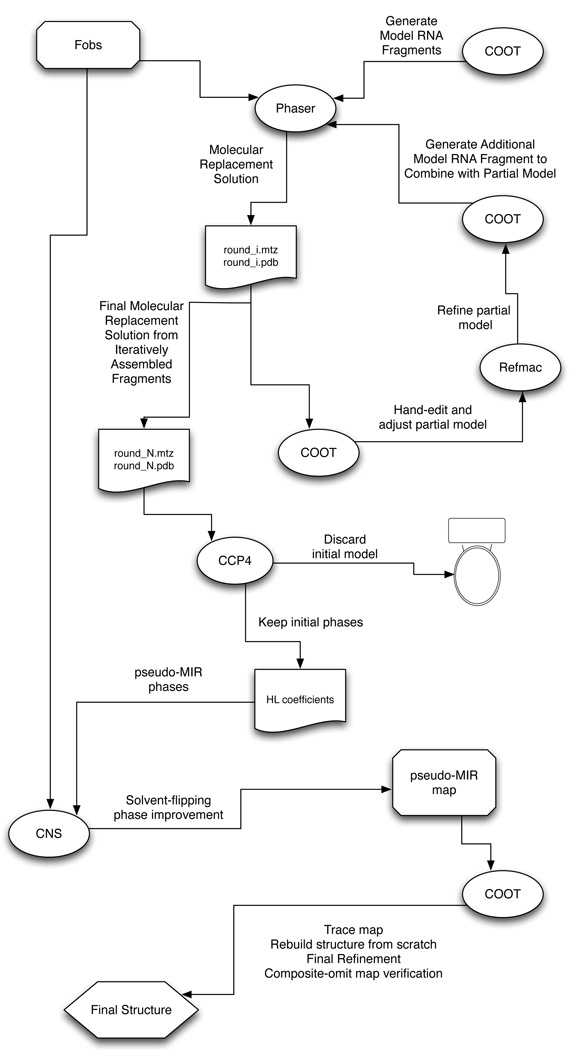
Figure 2. Schematic Representation of the Procedure.
An initial assembly of modeled RNA fragments (helices, loops) is used, in combination with a native data set (Fobs), to obtain a starting molecular replacement solution in PHASER [15] (round_1.mtz and round_1.pdb). This solution is examined in COOT [13,14], and all steric clashes are removed by manual editing, and any nucleotide that does not reside in strong electron density in the sigma-A-weighted 2Fo-Fc map is manually excised. The remaining model is then positionally refined using REFMAC [16] (which can be done within COOT) and any unoccupied density is modeled and refined in COOT. In addition, a new helical fragment (typically 5 base-pairs of A-from helix) for a subsequent iteration of molecular replacement is generated. Using the edited and refined model as a fixed partial model, the next (ith) round of molecular replacement in PHASER and subsequent editing and refinement is carried out with the new helical fragment. This cycle is repeated until no more helical fragments can be added. At this point, at the Nth cycle, the best molecular replacement solution (round_N.mtz and round_N.pdb) is used to generate a solvent-flattened electron density map in CNS [18]. The model (round_N.pdb) is discarded, and the calculated phase probability distributions are converted to Hendrickson-Lattmann coefficients within CCP4 [17] and are then imported into CNS, along with the native data set (Fobs), and treated as if they were experimentally determined MIR phases to be solvent-flattened. The resulting map is then used for building the final model from scratch (i.e., without reference to the discarded molecular replacement solution round_N.pdb) as if the map were derived from experimental MIR phases. The resulting structure is then checked against a composite-omit map generated in CNS in which 10% of the model is omitted from each element of the composite, and phases are regenerated from a standard simulated annealing procedure with a starting temperature of 4000K.