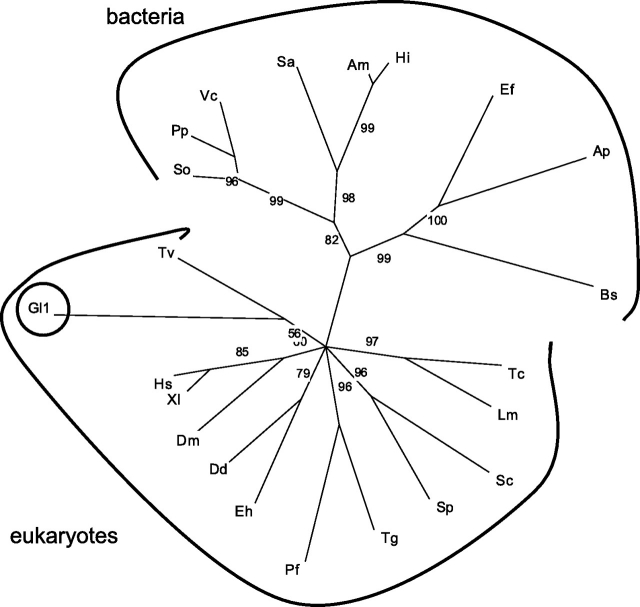
Fig. 3.
Phylogenetic analyses of phosphoglucomutases suggest that the Giardia PGM gene is deeply divergent. In the phylogenetic tree, which was constructed by maximum likelihood methods, the branch lengths are proportionate to differences between sequences, while the nodes indicate bootstrap support. Nodes with <50% bootstrap support were collapsed. The tree includes just one of the predicted PGMs of Giardia (Gl-PGM1) because the other Gl-PGM2 aligns so poorly with the rest of the PGMs. Representative protists include Dictyostelium discoideum (Dd), Entamoeba histolytica (Eh), Leishmania major (Lm), Plasmodium falciparum (Pf), Toxoplasma gondii (Tg), Trichomonas (Tv) and Trypanosoma cruzi (Tc). Other representative eukaryotes include Drosophila melanogaster (Dm), Homo sapiens (Hs), Schizosaccharomyces pombe (Sp), Saccharomyces cerevisiae (Sc) and Xenopus laevis (Xl). Representative eubacteria include Actinobacillus minor (Am), Anaerococcus prevotii (Ap), Borrelia spielmanii (Bs), Enterococcus faecalis (Ef), Haemophilus influenzae (Hi), Photobacterium profundum (Pp), Shewanella oneidensis (So), Streptomyces avermitilis (Sa) and Vibrio cholerae (Vc). No archaeal ortholog was identified