Abstract
Metabolomics is a rapidly evolving discipline that involves the systematic study of endogenous small molecules that characterize the metabolic pathways of biological systems. The study of metabolism at a global level has the potential to contribute significantly to biomedical research, clinical medical practice, as well as drug discovery. In this paper, we present the most up-to-date metabolite and metabolic pathway resources, and we summarize the statistical, and machine-learning tools used for the analysis of data from clinical metabolomics. Through specific applications on cancer, diabetes, neurological and other diseases, we demonstrate how these tools can facilitate diagnosis and identification of potential biomarkers for use within disease diagnosis. Additionally, we discuss the increasing importance of the integration of metabolomics data in drug discovery. On a case-study based on the Human Metabolome Database (HMDB) and the Chinese Natural Product Database (CNPD), we demonstrate the close relatedness of the two data sets of compounds, and we further illustrate how structural similarity with human metabolites could assist in the design of novel pharmaceuticals and the elucidation of the molecular mechanisms of medicinal plants.
1. Introduction
Metabolomics is a new technology that applies advanced separation and detection methods to capture the collection of small molecules that characterize metabolic pathways. This rapidly developing discipline involves the study of the total repertoire of small molecules present in the biological samples, particularly urine, saliva, and blood plasma [1]. Metabolites are the byproducts of metabolism, which is itself the process of converting food energy to mechanical energy or heat. Experts believe there are at least 3,000 metabolites that are essential for normal growth and development (primary metabolites) and thousands more unidentified (around 20,000, compared to an estimated 30,000 genes and 100,000 proteins) that are not essential for growth and development (secondary metabolites) but could represent prognostic, diagnostic, and surrogate markers for a disease state and a deeper understanding of mechanisms of disease [2]. Of particular interest to metabolomics researchers are small, low-molecular weight compounds that serve as substrates and products in various metabolic pathways [3].
Metabolomics, the study of metabolism at the global level, has the potential to contribute significantly to biomedical research, and ultimately to clinical medical practice [4, 5]. It is a close counterpart to the genome, the transcriptome and the proteome. Metabolomics, genomics, proteomics, and other “-omics” grew out of the Human Genome Project, a massive research effort that began in the mid-1990s and culminated in 2003 with a complete mapping of all the genes in the human body. When discussing the clinical advantages of metabolomics, scientists point to the “real-world” assessment of patient physiology that the metabolome provides since it can be regarded as the end-point of the “-omics” cascade [6]. Other functional genomics technologies do not necessarily predict drug effects, toxicological response, or disease states at the phenotype but merely indicate the potential cause for phenotypical response. Metabolomics can bridge this information gap since the identification and measurement of metabolite profile dynamics of host changes provides the closest link to the various phenotypic responses [7–9]. Thus it is clear that the global mapping of metabolic signatures pre- and postdrug treatment is a promising approach to identify possible functional relationships between medication and medical phenotype [10–13].
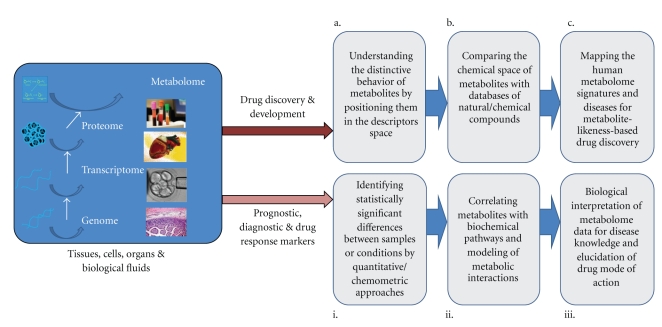
At the center of metabolomics is the concept that an individual's metabolite state is a close representation of the individuals overall health status. This metabolic state reflects what has been encoded by the genome and modified by environmental factors. In this paper, we demonstrate the enormous potential of metabolomics in disease monitoring and identification of prognostic, diagnostic, and drug response markers (Figure 1 (i)–(iii)), as well as in drug discovery and development in combination with systems chemical biology and chemoinformatics (Figures 1(a)–1(c)).
Figure 1.
Metabolomics holds the promise to deliver valuable information about biochemical pathways perturbed in disease and upon treatment, to monitor healthy people to detect early signs of disease, to diagnose disease or predict the risk of a disease, to subclassify disease, to make safer drugs by predicting the potential for adverse drug reactions, and to speed the discovery and development of novel drug molecules.
2. Databases and Data Analysis Tools
Databases of metabolites and metabolic reactions offer a wealth of information regarding the interaction of small molecules with biological systems, notably in relation with their chemical reactivity. In Table 1, we summarize all such metabolite and metabolic pathway resources which contain hundreds of reactions, metabolites, and pathways for several organisms and are designed to facilitate the exploration of metabolism across many different species. For example, the BiGG database (http://bigg.ucsd.edu/) is a metabolic reconstruction of human metabolism designed for systems biology simulation and metabolic flux balance modelling. It is a comprehensive literature-based genome-scale metabolic reconstruction that accounts for the functions of 1,496 ORFs, 2,004 proteins, 2,766 metabolites, and 3,311 metabolic and transport reactions. MassBank (http://www.massbank.jp/) is a mass spectral database of experimentally acquired high resolution MS spectra of metabolites. Maintained and supported by the JST-BIRD project, it offers various query methods for standard spectra obtained from Keio University, RIKEN PSC, and other Japanese research institutions. It is officially sanctioned by the Mass Spectrometry Society of Japan. The database has very detailed MS data and excellent spectral/structure searching utilities. More than 13,000 spectra from 1900 different compounds are available. The METLIN Metabolite Database (http://metlin.scripps.edu/index.jp) is a repository for mass spectral metabolite data. All metabolites are neutral or free acids. It is a collaborative effort between the Siuzdak and Abagyan groups and Center for Mass Spectrometry at The Scripps Research Institute. METLIN is searchable by compound name, mass, formula, or structure. It contains 15,000 structures, including more than 8000 di- and tripeptides. METLIN contains MS/MS, LC/MS and FTMS data that can be searched by peak lists, mass range, biological source or disease. Below we describe in more detail three interconnected databases; the Human Metabolome Database (http://www.hmdb.ca/), the Small Molecule Pathway Database (http://www.smpdb.ca/) and the Toxin and Toxin-Target Database (www.t3db.org/) (Figure 2).
Table 1.
Machine-learning algorithms often used in metabolomics.
| Technique | Description |
|---|---|
| PCA | The Principal Component Analysis (PCA) is a frequently used method which is applied to extract the systematic variance in a data matrix. It helps to obtain an overview over dominant patterns and major trends in the data. The aim of PCA is to create a set of latent variables which is smaller than the set of original variables but still explains all the variance of the original variables. In mathematical terms, PCA transforms a number of correlated variables into a smaller number of uncorrelated variables, the so-called principal components. |
| PLS | Partial Least Squares (PLS), also called Projection to Latent Structures, is a linear regression method that can be applied to establish a predictive model, even if the objects are highly correlated. The X variables (the predictors) are reduced to principal components, as are the Y variables (the dependents). The components of X are used to predict the scores on the Y components, and the predicted Y component scores are used to predict the actual values of the Y variables. In constructing the principal components of X, the PLS algorithm iteratively maximizes the strength of the relation of successive pairs of X and Y component scores by maximizing the covariance of each X-score with the Y variables. This strategy means that while the original X variables may be multicollinear, the X components used to predict Y will be orthogonal. Also, the X variables may have missing values, but there will be a computed score for every case on every X component. Finally, since only a few components (often two or three) will be used in predictions, PLS coefficients may be computed even when there may have been more original X variables than observations. |
| O-PLS | The Orthogonal Projections to Latent Structures (O-PLS) is a linear regression method similar to PLS. However, the interpretation of the models is improved because the structured noise is modeled separately from the variation common to X and Y. Therefore, the O-PLS loading and regression coefficients allow for a more realistic interpretation than PLS, which models the structured noise together with the correlated variation between X and Y. Furthermore, the orthogonal loading matrices provide the opportunity to interpret the structured noise. |
| PLS-DA | PLS-Discriminant Analysis (PLS-DA) is a frequently used classification method that is based on the PLS approach, in which the dependent variable is chosen to represent the class membership. PLS-DA makes it possible to accomplish a rotation of the projection to give latent variables that focus on class separation. The objective of PLS-DA is to find a model that separates classes of objects on the basis of their X-variables. This model is developed from the training set of objects of known class membership. The X-matrix consists of the multivariate characterization data of the objects. To encode a class identity, one uses as Y-data a matrix of dummy variables, which describe the class membership. A dummy variable is an artificial variable that assumes a discrete numerical value in the class description. The dummy matrix Y has G collumns (for G classes) with ones and zeros, such that the entry in the gth column is one and the entries in other columns are zero for observations of class g. |
| ANN | Artificial Neural Networks (ANN) is a method, or more precisely a set of methods, based on a system of simple identical mathematical functions, that working in parallel yield for each multivariate input X a single or multiresponse answer. ANN methods can only be used if a comparably large set of multivariate data is available which enables ANN training by example and work best if they are dealing with nonlinear relationships between complex inputs and outputs. The main component of a neural network is the neuron. Each neuron has an activation threshold, and a series of weighted connections to other neurons. If the aggregate activation a neuron receives from the neurons connected to it exceeds its activation threshold, the neuron fires and relays its activation to the neurons to which it is connected. The weights associated with these connections can be modified by training the network to perform a certain task. This modification accounts for learning. ANN are often organized into layers, with each layer receiving input from one adjacent layer, and sending it to another. Layers are categorized as input layers, output layers, and hidden layers. The input layer is initialized to a certain set of values, and the computations performed by the hidden layers update the values of the output layers, which comprise the output of the whole network. Learning is accomplished by updating the weights between connected neurons. The most common method for training neural networks is back propagation, a statistical method for updating weights based on how far their output is from the desired output. To search for the optimal set of weights, various algorithms can be used. The most common is gradient descent, which is an optimization method that, at each step, searches in the direction that appears to come nearest to the goal. |
| SOM | Self-Organizing Maps (SOM) or Kohonen network is an unsupervised neural network method which has both clustering and visualization properties. It can be used to classify a set of input vectors according to their similarity. The result of such a network is usually a two-dimensional map. Thus, SOM is a method for projecting objects from a high dimensional data space to a two-dimensional space This projection enables the input data to be partitioned into “similar” clusters while preserving their topology, that is, points that are close to one another in the multidimensional space are neighbors in the two-dimensional space as well. |
| SVM | Support Vector Machines (SVM) perform classification by constructing an N-dimensional hyperplane that optimally separates the data into two categories. A SVM model using a sigmoid kernel function is equivalent to a two-layer, perceptron neural network. The task of choosing the most suitable representation is known as feature selection. A set of features that describes one object is called a vector. The goal of SVM modeling is to find the optimal hyperplane that separates clusters of vectors in such a way that objects with one category of the target variable are on one side of the plane and objects with the other category are on the other size of the plane. The vectors near the hyperplane are the support vectors. |
| K-means | K-means is a classic clustering technique that aims to partition objects into k clusters. First, you specify k, that is, how many clusters are being sought. Then, k points are chosen at random as cluster centers. All objects are assigned to their closest cluster center according to the ordinary Euclidean distance metric. Next, the centroid, or mean, of the objects in each cluster is calculated. These centroids are taken to be the new center values for their, respective clusters. Finally, the whole process is repeated with the new cluster centers. Iteration continues until the same points are assigned to each cluster in consecutive rounds, at which stage the cluster centers have stabilized. |
| Genetic Algorithms | Genetic algorithms are nondeterministic stochastic search/optimization methods that utilize the evolutionary concepts of selection, recombination or crossover, and mutation into data processing to solve a complex problem dynamically. Possible solutions to the problem as so-called artificial chromosomes, which are changed and adapted throughout the optimization process until an optimus solution is obtained. A set of chromosomes is called population and creation of a population from a parent population is called generation. In a first step, the original population is created. For each chromosome, the fitness is determined and a selection algorithm is applied to choose chromosomes for mating. These chromosomes are then subject to the crossover, and the mutation operators, which finally yield a new generation of chromosomes. |
Figure 2.
A screenshot montage of the HMDB, SMPDB and T3DB databases.
2.1. Human Metabolome Database (HMDB)
Focusing on quantitative, analytic, or molecular scale information about metabolites, the enzymes and transporters associated with them, as well as disease related properties the HMDB represents the most complete bioinformatics and chemoinformatics medical information database. It contains records for thousands of endogenous metabolites identified by literature surveys (PubMed, OMIM, OMMBID, text books), data mining (KEGG, Metlin, BioCyc) or experimental analyses performed on urine, blood, and cerebrospinal fluid samples. The annotation effort is aided by chemical parameter calculators and protein annotation tools originally developed for DrugBank. The HMDB is fully searchable with many built-in tools for viewing, sorting, and extracting metabolites, biofluid concentrations, enzymes, genes, NMR or MS spectra and disease information. The HMDB currently contains 7,985 compounds that are linked to 69,295 different synonyms. These compounds are also connected to 908 C-NMR and 916 H-NMR spectra as well as 7,234 associated enzymes. All chemical structures in these pathway maps are hyperlinked to HMDB MetaboCards and all enzymes are hyperlinked to UniProt data cards for human enzymes. The majority of the compounds have been detected in blood (4,226) while 784 compounds were detected in urine, 363 in CSF (cerebrospinal fluid) and 315 in other biofluids. In order a compound to be included in the HMDB it must fulfil certain criteria; it should be of biological origin, the compound weight must be <1,500 Da, and it should be found at concentrations greater than 1 mM in one or more biofluids/tissues. Compounds that are not covered by the above description but are either biomedically important metabolites, like hormones, or certain very common drugs and some ubiquitous food additives, like vitamins, are some notable exceptions in the HMDB. For a large number of metabolites the concentration values in the biofluids are given with data for both normal and abnormal values.
A key feature that distinguishes the HMDB from other metabolic resources is its extensive support for higher level database searching and selecting functions. More than 175 hand-drawn-zoomable, fully hyperlinked human metabolic pathway maps can be found in HMDB and all these maps are quite specific to human metabolism and explicitly show the subcellular compartments where specific reactions are known to take place. As an equivalent to BLAST the HMDB contains a structure similarity search tool for chemical structures and users may sketch or paste a SMILES string of a query compound into the Chem-Query window. Submitting the query launches a structure similarity search tool that looks for common substructures from the query compound that match the HMDB's metabolite database. The wealth of information and especially the extensive linkage to metabolic diseases to normal and abnormal metabolite concentration ranges, to mutation/SNP data and to the genes, enzymes, reactions and pathways associated with many diseases of interest makes the HMDB one the most valuable tool in the hands of clinical chemists, nutritionists, physicians and medical geneticists.
2.2. The Small Molecule Pathway Database (SMPDB)
SMPDB consists of approximately 350 hand-drawn pathways with more than 280 of them unique to SMPDB. These pathways describe small molecule metabolism or small-molecule processes that are specific to humans and fall into four different categories: (i) metabolic pathways; (ii) small-molecule disease pathways, (iii) small molecule drug pathways, and (iv) small molecule signalling pathways. In order for a metabolic pathway to be suitable for inclusion in SMPDB, it must be found in humans and it must contain at least five small molecules. If it is a human disease, drug or signalling pathway the determining factor for inclusion is its central feature being based on the action of at least one small molecule. More specifically, in SMPDB, disease pathways refer to those pathways describing human disease processes where small-molecule metabolite dysregulation is the primary hallmark of the disease. For qualifying a small molecule or set of small molecules to be included in SMPDB, a significant concentration change, which is commonly used for the diagnosis, prognosis, or monitoring for a given disease, is required. The SMPDB interface is largely modelled after the interface used for DrugBank and the HMDB with a navigation panel for browsing, searching, and downloading the database. The users can choose between two browsing options, SMP-BROWSE, and SMP-TOC. The latter is basically a scrollable hyperlinked table of contents that lists all pathways by name and category. SMP-BROWSE is a more comprehensive browsing tool that provides a tabular synopsis of SMPDB's content using thumbnail images of the pathway diagrams, textual descriptions of the pathways, as well as lists of the corresponding chemical components and enzyme/protein components. All of the chemical structures and proteins/enzymes illustrated in SMPDB's diagrams are hyperlinked to other online databases or tables, but this is common in most pathway databases. Specifically, all metabolites, drugs or proteins shown in the SMP-BROWSE tables or in a pathway diagram are linked to HMDB, DrugBank or UniProt, respectively. One of the most interesting search options in SMPDB is the SMP-MAP which offers both multiidentifier searches as well as transcriptomic, proteomic, or metabolomic mapping. SMP-MAP allows users to select the type of “-omic” data, then paste in a list of identifiers and have a table generated of appropriately highlighted pathways containing those components.
The content of SMPDB is not normally found in other pathway databases with 281 unique pathways (in total of 364). More specifically, 154/168 drug pathways, 11/13 metabolite signalling pathways, 4/70 metabolic pathways and 112/113 metabolic disease pathways of the SMPDB cannot be found in any of the known databases (KEGG, Reactome, EHMN, WikiPathways, HumanCyc, BioCarta, and PharmGKB). Especially in relation to metabolic disease pathways and drug pathways the SMPDB is currently the only pathway database that includes significant numbers of them. In addition SMPDB offers a significant amount of useful graphical content including the depiction of the relevant organs, cellular locations, organelles, cofactors and other cellular features. Because SMPDB is focused on small molecules, it does not include the key protein signalling pathway information which limits significantly its use in comparative metabolic studies, protein network analysis, metabolic engineering or metabolic evolution.
2.3. Toxin and Toxin-Target Database (T3DB)
As the name indicates, T3DB is primarily intended to be a database that links toxins with their biological targets. However, the molecular interaction information is further supplemented with detailed descriptions of the toxin's mechanism of action, its metabolism in the human body, its lethal or toxic dose levels, its potential carcinogenicity, exposure sources, symptoms or health effects and suggested treatment options. More than 2,900 toxin entries corresponding to more than 34,000 different synonyms are currently included in the T3DB. T3DB toxins were identified using a number of methods that include data mining, literature surveys, toxicology textbooks but also examining lists of controlled or banned substances. The toxic compounds that were identified were subsequently used to derive additional substances that were toxic by relation. In order to ensure both completeness and correctness each toxin record entered in T3DB was reviewed by two different members of the team. Much of the annotation was done manually especially in areas such as route of delivery, mechanisms of action, health effects and target identification.
T3DB contains compounds that have been routinely identified as hazardous in relatively low concentrations (<1 mM for some, <1 μM for others) and which appear on multiple toxin/poison lists provided by government agencies such as TOXNET or the toxicological and medical literature. In each case, the toxicity of each compound was assessed by examining the available toxicity measurements and health effects, such as minimum lethal dose, LD50, LC50 values and carcinogenicity. In addition these toxins are further connected to approximately 1,300 protein targets through almost 33,500 toxin and toxin-target bonds. All the above information is supported by more than 3,100 references. To facilitate browsing, the T3DB is divided into synoptic summary tables which, in turn, are linked to more detailed “Tox-Cards”-in analogy to the very successful “DrugCard” concept found in DrugBank. Each Tox-Card entry contains over 80 data fields, with ~50 data fields devoted to chemical and toxicological/medical data and ~30 data fields devoted to describing the toxin target(s). In addition to the data viewing and sorting features, the T3DB also offers a local BLAST search that supports both single and multiple sequence queries, a boolean text search based on KinoSearch (http://www.rectangular.com/kinosearch/), a chemical structure utility based on ChemAxon's MarvinView, and a relational data extraction tool similar to that found in DrugBank and the HMDB. The SeqSearch, a sequence searching utility of T3DB's, provides the option to search through T3DB's collection of 1,300 known human toxin targets. The SeqSearch makes possible the identification of both orthologous and paralogous targets for known toxins or toxin targets but facilitates also the identification of potential targets of other animal species. The T3DB's data extraction utility employs a simple relational database system that allows users to select one or more data fields and to search for ranges, occurrences or partial occurrences of words, strings or numbers.
In comparison to other databases that contain toxic substances T3DP probably has the smallest number of toxins or poisons in its collection since T3DB was designed as a database for common toxins and not for all known toxic substances. A key focus of the T3DB is on providing “depth” over “breath” and with its unique emphasis on “common” substances should prove to be a valuable resource in toxicometabolomics and clinical toxicology research.
3. Identification of Disease Biomarkers
In clinical metabolomics one is almost always working with a biofluid or a fluidized tissue extract. The preference of working with biofluids over tissues is primarily dictated by the fact that fluids are far easier to process and analyze. Likewise the collection of biofluids is generally much less invasive than the collection of tissues. Biofluids analysis is always done with the assumption that the chemicals found in different biofluids are largely reflective of the biological state of the organ that produces or is bathed in this fluid. Metabolomics share many of the computational needs with genomics, proteomics, and transcriptomics. All four “-omics” techniques require electronically accessible and searchable databases, all of them require software to handle or process data from their own high-throughput instruments, all of them require laboratory information systems to manage their data and all require software tools to predict or model properties, pathways, relationships, and processes [14]. In terms of data analysis, metabolomics, like other functional genomics technologies, produces high-dimensional datasets, and so it is amenable to many of the analyses applied to microarray data. Statistical modelling (Table 2) range from univariate statistical testing to multivariate regression methods such as principal component analysis (PCA), partial least squares (PLS) or orthogonal projections to latent squares (OPLS), cluster analysis, machine-learning techniques and nonlinear methods, for example Kohonen's self organizing maps (SOM), support vector machines (SVM), and neural networks (NN) [15–18]. In the following section we have chosen to focus on specific applications that demonstrate how the above statistical modelling tools can facilitate the diagnosis of diseases and the identification of potential biomarkers for use within disease diagnosis.
Table 2.
Freely available databases on metabolic pathways and the metabolome.
| Metabolic Pathways Databases | Webpage |
|---|---|
| BRENDA, the enzyme database, has comprehensive information on enzymes and enzymatic reactions. It is one of several databases nested within the metabolic pathway database set of the SRS5 sequence retreival system at EBI. | http://www.brenda.uni-koeln.de/ |
| Reactome is an online bioinformatics database of biology described in molecular terms. The largest set of entries refers to human biology, but Reactome covers a number of other organisms as well. It is an on-line encyclopedia of core human pathways-DNA replication, transcription, translation, the cell cycle, metabolism, and signaling cascades. | http://www.reactome.org/ |
| KEGG Metabolic Pathways include graphical pathway maps for all known metabolic pathways from various organisms. Ortholog group tables, containing conserved, functional units in a molecular pathway or or assembly, as well as comparative lists of genes for a given functional unit in different organisms, are also available. | http://www.genome.jp/kegg/metabolism.html |
| MetaCyc is a database of nonredundant, experimentally elucidated metabolic pathways. MetaCyc contains more than 1,400 pathways from more than 1,800 different organisms, and is curated from the scientific experimental literature. MetaCyc contains pathways involved in both primary and secondary metabolism, as well as associated compounds, enzymes, and genes. | http://metacyc.org/ |
| The WIT Metabolic Reconstruction project produces metabolic reconstructions for sequenced, or partially sequenced, genomes. It currently provides a set of over 25 such reconstructions in varying states of completion. Over 2900 pathway diagrams are available, associated with functional roles and linked to ORFs. | http://ergo.integratedgenomics.com/ |
| BioCarta website provides gene interactions in dynamic graphical models. The online maps depicts molecular relationships and it catalogs and summarizes important resources providing information for more than 12,000 genes from multiple species. It contains both classical pathways as well as suggestions for new pathways. | http://main.biocarta.com/genes/index.asp |
| EcoCyc describes the genome and the biochemical machinery of E. coli. It provides a molecular and functional catalog of the E. coli cell to facilitates system-level understanding. Its Pathway/Genome Navigator user interface visualizes the layout of genes, of individual biochemical reactions, or of complete pathways. It also supports computational studies of the metabolism, such as pathway design, evolutionary studies, and simulations. A related metabolic database is Metalgen. | http://ecocyc.org/ |
| BioSilico is a web-based database system that facilitates the search and analysis of metabolic pathways. Heterogeneous metabolic databases including LIGAND, ENZYME, EcoCyc and MetaCyc are integrated in a systematic way, thereby allowing users to efficiently retrieve the relevant information on enzymes, biochemical compounds and reactions. In addition, it provides well-designed view pages for more detailed summary information. | http://mbel.kaist.ac.kr/lab/index_ko.html |
| EXPASY - Biochemical Pathways is a searchable database of metabolic pathways, enzymes, substrates and products. Based on a given search, it produces a graphic representation of the relevant pathway(s) within the context of an enormous metabolic map. Neighboring metabolic reactions can then be viewed through links to adjacent maps. | http://www.expasy.ch/cgi-bin/search-biochem-index |
| BioPath is a database of biochemical pathways that provides access to metabolic transformations and cellular regulations derived from the Roche Applied Science “Biochemical Pathways” wall chart. BioPath provides access to biological transformations and regulations as described on the “Biochemical Pathways” chart. | http://www.molecular-networks.com/biopath/ |
| BioCyc is a collection of 505 Pathway/Genome Databases. Each database in the BioCyc collection describes the genome and metabolic pathways of a single organism. The BioCyc Web site contains many tools for navigating and analyzing these databases, and for analyzing omics data, including the following: Genome browser, Display of individual metabolic pathways, and of full metabolic maps, Visual analysis of user-supplied omics datasets by painting onto metabolic map, regulatory map, and genome map, Comparative analysis tools. | http://biocyc.org/ |
| Metabolome Databases | Webpage |
| The Biological Magnetic Resonance Data Bank (BMRB) focuses on quantitative data generated by spectroscopic investigations of biological macromolecules. It has links to search engines such as PubChem, that connect to recent articles and new data. It also links to projects and other databases that are all related to Metabolomics and Metabonomics. This database focuses on the NMR research aspect of metabolites discovery and their role in metabolism. | http://www.bmrb.wisc.edu/metabolomics/ |
| The Madison Metabolomics Consortium Database contains metabolites determined through NMR and MS. It contains information with the main focus on Arabidopsis thaliana, but also refers to many different species. The database also contains information on the presence of metabolites under several different physiological conditions, their structures in 2D and 3D, and links to related resource sources and other databases. | http://mmcd.nmrfam.wisc.edu/ |
| The Human Metabolome Database is an extremely comprehensive, free electronic database that gives a detailed overview of human metabolites divided into chemical, clinical, and molecular biology/biochemistry data. | http://www.hmdb.ca/ |
| KNApSAcK is a Java application that presents an interactive display of biochemical information that can be searched by organism or metabolite name. KNApSAcK focuses primarily on the origin and mass spectra of particular metabolites. | http://kanaya.naist.jp/KNApSAcK |
| The BiGG database is a metabolic reconstruction of human metabolism designed for systems biology simulation and metabolic flux balance modeling. It is a comprehensive literature-based genome-scale metabolic reconstruction that accounts for the functions of 1,496 ORFs, 2,004 proteins, 2,766 metabolites, and 3,311 metabolic and transport reactions. It was assembled from build 35 of the human genome. | http://bigg.ucsd.edu/ |
| SetupX, developed by the Fiehn laboratory at UC Davis, is a web-based metabolomics LIMS. It is XML compatible and built around a relational database management core. It is particularly oriented towards the capture and display of GC-MS metabolomic data through its metabolic annotation database called BinBase. | http://fiehnlab.ucdavis.edu:8080/m1/ |
| McGill-MD is a metabolome database containing metabolite mass spectra of organisms; with abiotic/biotic stress or in homeostasis. Users are able to obtain a table containing the metabolome of an organism, or download mass spectra of all the metabolites entered in the database. | http://metabolomics.mcgill.ca/ |
| SYSTOMONAS (SYSTems biology of pseudOMONAS) is a database for systems biology studies of Pseudomonas species. It contains extensive transcriptomic, proteomic and metabolomic data as well as metabolic reconstructions of this pathogen. Reconstruction of metabolic networks in SYSTOMONAS was achieved via comparative genomics. Broad data integration with well established databases BRENDA, KEGG and PRODORIC is also maintained. | http://www.systomonas.de/ |
| MassBank is a mass spectral database of experimentally acquired high resolution MS spectra of metabolites. Maintained and supported by the JST-BIRD project, it offers various query methods for standard spectra obtained from Keio University, RIKEN PSC, and other Japanese research institutions. It is officially sanctioned by the Mass Spectrometry Society of Japan. The database has very detailed MS data and excellent spectral/structure searching utilities. More than 13,000 spectra from 1900 different compounds are available. | http://www.massbank.jp/ |
| The Golm Metabolome Database provides public access to custom GC/MS libraries which are stored as Mass Spectral (MS) and Retention Time Index (RI) Libraries (MSRI). These libraries of mass spectral and retention time indices can be used with the NIST/AMDIS software to identify metabolites according their spectral tags and RI's. The libraries are both searchable and downloadable and have been carefully collected under defined conditions on several types of GC/MS instruments (quadrupole and TOF). | http://csbdb.mpimp-golm.mpg.de/csbdb/gmd/gmd.html |
| The METLIN Metabolite Database is a repository for mass spectral metabolite data. All metabolites are neutral or free acids. It is a collaborative effort between the Siuzdak and Abagyan groups and Center for Mass Spectrometry at The Scripps Research Institute. METLIN is searchable by compound name, mass, formula or structure. It contains 15,000 structures, including more than 8000 di and tripeptides. METLIN contains MS/MS, LC/MS and FTMS data that can be searched by peak lists, mass range, biological source and or disease. | http://metlin.scripps.edu/index.php |
3.1. Cancer
The paper of Guan et al. [19] is the first application of SVMs and SVM-related feature selection methods (recursive feature elimination with linear and nonlinear kernel, L1SVM, and Weston's method) for classifying LC/TOF MS data of serum samples from ovarian cancer patients and control. Sera from 37 ovarian cancer patients and 35 benign controls were studied and three evaluation processes (leave-one-out-cross-validation, 12-fold-cross-validation, and 52–20-split-validation) were used to examine the SVM models based on selected potential metabolic diagnostic biomarkers in terms of their ability for differentiating control versus disease serum samples. Classification of the serum sample test set was over 90% accurate indicating promise that this approach may lead to the development of an accurate and reliable metabolomic-based protocol for detecting ovarian cancer.
The aim of another recent study [20] was to elucidate the predictability of breast cancer by means of urinary excreted nucleosides. The authors analyzed a balanced set of 170 urine samples, 85 breast cancer women and, respective healthy controls, and after identification of 51 nucleosides/ribosylated metabolites in the urine of breast cancer women a valid set of 35 candidates was selected for subsequent computational analysis. The bioinformatic tool of Oscillating Search Algorithm for Feature Selection (OSAF) was applied to iteratively improve features for training of SVMs to better predict breast cancer. The authors found a reasonable set of tumor-related metabolite pairs with SVM prediction performance of 83.5% sensitivity and 90.6% specificity, demonstrating that semiquantitative measurements are valuable for pattern detection using nonparametric machine-learning algorithms.
Arakaki et al. [21] described CoMet, a fully automated and general computational metabolomics method that uses a Systems Biology approach to predict the human metabolites which intracellular levels are more likely to be altered in cancer cells. The authors then prioritize the metabolites predicted to be lowered in cancer compared to normal cells as potential anticancer agents. They discovered eleven metabolites that either alone or in combination exhibit significant antiproliferative activity in Jurkat leukemia cells. Nine of these metabolites that were predicted to be lowered in Jurkat cells with respect to lymphoblasts (riboflavin, tryptamine, 3-sulfino-L-alanine, menaquinone, dehydroepiandrosterone, α-hydroxystearic acid, hydroxyacetone, seleno-L-methionine and 5,6-dimethylbenzimidazole) exhibited antiproliferative activity that has not been reported before. These results strongly suggest that many other metabolites with important roles in cellular growth control may be waiting to be discovered, opening up the possibility of novel approaches against cancer. CoMet adopts the viewpoint that the cell is an integrated machine and the author's resulting simple hypothesis that inspired its creation can greatly assist in the understanding of the contribution of metabolism to this complex disease.
In a different approach using an animal model Southam et al. [22] applied NMR-based metabolomics to histpathologically well-characterized livers dissected from a wild-caught species of marine flatfish. The use of metabolic profiling and correlation networks enabled a more thorough interpretation of this dataset. Fingerprint analysis identified single metabolites that showed concentration changes between phenotypes, while network analysis highlighted alterations to the relationships of paired metabolites between phenotypes. Tumor tissues showed elevated anaerobic respiration and reduced TCA cycle activity, while alanine and proline were indicated to supplement pyruvate (and NAD+) production during anaerobic metabolism in the tumor tissue. Choline metabolism was altered in tumor including disruptions of the choline oxidation and CDP-choline pathways. The author's hypothesis was that such disruption of the choline oxidation pathway could lead to reduced SAM production and potentially DNA hypomethylation of oncogenes.
3.2. Diabetes
The paper of Altmaier et al. [23] presents a bioinformatics analysis of what can be considered as a standard experimental setting of a preclinical drug testing experiment with two independent factors, “state” and “medication”. Targeted quantitative metabolomics covering a wide range of more than 800 relevant metabolites were measured in blood plasma samples from healthy and diabetic mice under rosiglitazone (a member of thiazolidinedione) treatment. The authors show that known and new metabolic phenotypes of diabetes and medication can be recovered in a statistically objective manner. Analyzing ratios between metabolite concentrations dramatically reduces the noise in the data set allowing the discovery of new potential biomarkers of diabetes, such as the N-hydroxyacyloylsphingosyl-phosphocholines SM(OH)28 : 0 and SM(OH)26 : 0. Using a hierarchical clustering technique on partial η2 values the authors identified functionally related groups of metabolites, indicating a diabetes-related shift from lysophosphophatidylcholine to phosphatidylcholine levels.
Coupled LC/MS technology to multivariate statistical analysis in order to study phospholipid metabolic profiling in diabetes mellitus and to discover the potential biomarkers was the approach of Wang et al. [24]. PCA and PLS-DA models were compared in class separation of type 2 diabetes mellitus (DM2) patients and healthy controls. Uv (unit variance) scaling and OSC (orthogonal signal correction) data preprocessing methods were also developed to improve class separation. Using the supervised PLS-DA algorithm with Uv scaling and OSC technique on the data set, it was found that the separation of different classes was highly improved (compared to PCA analysis) particularly with OSC. The application of LC/MS coupled to PLS-DA of data with OSC scaling made it possible to classify DM2 and control and further to discover potential biomarkers that can be identified by MS/MS.
NMR-based metabolomics coupled with sophisticated bioinformatics was shown capable of identifying rapid changes in global metabolite profiles in urine and plasma (treatment “fingerprints”) which may be linked to the well-documented early changes in hepatic insulin sensitivity following thiazolidinedione intervention in Type 2 diabetes mellitus [12]. Several endogenous metabolites in urine and plasma of T2DM patients that responded to rosiglitazone treatment were identified. In urine these changes were related to a gender-independent relative reduction of hippurate and a further increase of aromatic acids. The gender-dependent changes observed in plasma samples included an increase in branched chain amino acids, alanine, glutamine/glutamate and citrate, coinsiding with a decrease in lactate, acetate, tyrosine, and phenyalanine in the female T2DM group, where changes in the male T2DM group included an increase in branched chain amino acids, alanine, glutamine, and threonine. A good distinction between diabetic patients and healthy volunteers as well as separation by gender was accomplished when Supervised Principal Component Discriminant Analysis (PC-DA) of plasma or urine samples was applied which comprises an important new addition to the early clinical development “proof of concept” toolbox for thiazolidinediones.
Diabetes is associated with increased incidence of vascular complications, and premature aging. In the study of Makinen et al. [25], the emphasis was on the metabolic continuum that underlies the slow and often elusive development of chronic complications. The authors obtained serum samples to measure two molecular windows, —the lipoprotein lipids (LIPO) window and the low molecular weight molecules (LMWM)—for 613 patients with type I diabetes, and diverse spread of complications. The H-NMR analyses combined with SOM instead of linear decomposition methods allowed the authors to transform the spectral data into an accesible form of information. The work of Makinen et al. demonstrated the limitations of single diagnostic biomarkers and illustrated a fundamental diagnostic challenge. Even though there is a common biochemical basis of diabetic kidney disease, diabetic retinal disease, the metabolic syndrome, and macrovascular diseases however they do not conclusively define each other.
Salek et al. [26] describe the application of 1H-NMR spectroscopybased metabolomics, combined with multivariate and univariate statistics, to investigate the urinary metabolic profiles in two animal models (mice and rat) of T2DM, and they compared these metabolic changes with perturbations observed in a human population. This study demonstrated metabolic similarities between the three species examined. Along with the expected changes in hepatic glycolysis/ gluconeogenesis changes in the excretion of TCA cycle intermediates, polyols, amines, and amino acids were detected. Furthermore significant changes in pyruvate and fatty acid metabolism as well as hepatic amino acid metabolism were observed including tryptophan metabolism. A profound perturbation in nucleotide metabolism, previously linked with peroxisome proliferation, was also observed and may indicate a metabolic consequence of substrate excess in many tissues, especially the liver.
In the study of Connor et al. [27], the authors have generated NMR-based metabolomic and transcriptomic data from the db/db diabetic mouse, one of the most extensively studied animal models of T2D. Db/db mice lack a functioning leptin receptor resulting in defective leptin-mediated signal transduction. Metabolomics data identified 24 distinct pathways that were altered in the diabetic mice when compared to their euglycaemic littermates. Several of these pathways were related to known disease effects, but in addition novel effects on branched chain amino acid metabolism, nicotinamide metabolites, pantothenic acid, and gut microflora metabolism were also observed. Integrative pathway analysis of the metabolite-centric networks and the cross-platform transcriptomics and metabolomics results effectively linked many of the metabolite changes to pathways involved in gluconeogenesis, and those generating substrates for gluconeogenesis, mitochondrial dysfunction and oxidative stress, and altered protein turnover. Overall, these metabolites are likely reflective of additional underlying pathophysiology that is present in T2D.
The objective of Lanza et al. [28] was to illustrate the utility of a combination of analytical methods and multivariate statistical analysis for detecting a metabolic fingerprint that reflects known pathways that are altered with insulin deficiency. The authors analyzed plasma from type 1 diabetic (T1D) humans during insulin treatment (I+) and acute insulin deprivation (I−) and nondiabetic participants (ND) and they generated correlation matrices for the plasma metabolites measured by both MS and NMR to create a compendium metabolic profile that integrates the complementary information from the two analytical methods. Multivariate statistics differentiated proton spectra from I− and I+ based on several derived plasma metabolites that were elevated during insulin deprivation (lactate, acetate, allantoin, and ketones) as well as several underlying physiological processes that are known to be altered by short-term insulin deprivation in type 1 diabetic people (e.g., mitochondrial dysfunction, oxidative stress, protein synthesis, degradation, and oxidation, gluconeogenesis, and ketogenesis).
Bao et al. [29] performed a metabonomic study to determine metabolic variations associated with T2DM and the drug treatments on 74 patients who were newly diagnosed with T2DM and received a 48-week treatment of a single drug, repaglinide, metformin, or rosiglitazone. A total of 212 individual metabolites were consistently detected in at least 90% of the serum samples and orthogonal projections to latent structures discriminant analysis, a newly developed supervised pattern recognition method, was used to capture the subtle intergroup variations and establish a prediction model to assess the physiological impact by drug treatment. As compared to healthy controls, the altered serum metabolites in diabetic subjects, include the significantly increased valine, maltose, glutamate, urate, butanoate, and long-chain fatty acid (C16:0, C18:1, C18:0, octadecanoate, and arachidonate), and decreased glucuronolactone, lysine, and lactate suggesting a hypercatabolic state in T2DM patients. Rosiglitazone treatment was able to reverse more abnormally expressed metabolites, such as valine, lysine, glucuronolactone, C16:0, C18:1, urate, and octadecanoate, than the other two drugs.
Čuperlović-Culf [30] presented an application of fuzzy K-means (F-KM) method for the classification of metabolic profiles of urine samples in diabetic patients. F-KM is a fuzzy version of standard K-means clustering. In F-KM clustering, each sample has an overall membership, that is, sum of membership values for all clusters, of 1. This overall membership is appointed to clusters based on the similarity between the sample's metabolic fingerprint and the profile of cluster's centroid. From the membership values, it is then possible to determine different levels of coclustering between samples-based on the top membership, second highest membership, and so forth. In their work different clustering methods were compared with F-KM. For human type II diabetes and healthy phenotypes membership values, F-KM lead to better sample separation while it was the only method that allowed distinction on both major groups and sample subtypes.
3.3. Neurological and Other Diseases
The study of Rozen et al. [4] was designed to assess whether there are systematic differences between redox-active metabolites in the blood of patients with motor neuron disease (MND) and healthy controls by analyzing the blood plasma of 30 healthy controls and 28 individuals with MND. To determine which metabolites were significantly elevated or reduced in MND the authors used three measures of class association, the t-statistic, Pearson's correlation coefficient, and the “relative class association” measure. All three measures produced similar rankings of their metabolites by their level of association with MND versus control. The authors assessed statistical significance by permutation testing and all measures showed similar numbers of metabolites to have significantly higher or lower concentrations in MND compared to controls. Subsequently they analyzed these data to determine if the metabolites were capable of distinguishing four subgroups (normal controls MND patients taking riluzole medication, MND without riluzole medication, and the subgroup enriched for LMN-lower motor neuron disease) using the 317 metabolite concentrations. Using PLS-DA, a supervised projection technique, the authors found a three-dimensional projection in which these four subgroups were significantly separated.
1H nuclear magnetic resonance spectroscopy in conjunction with computerized pattern recognition analysis were employed to investigate metabolic profiles of a total of 152 cerebrospinal fluid (CSF) samples from drug-naïve or minimally treated patients with first-onset paranoid schizophrenia and healthy controls [7]. Plots of PLS-DA scores showed a clear differentiation between healthy volunteers and drug-naïve patients. The PLS-DA score plots show that atypical antipsychotic drug treatment results in a shift of approximately 50% of patients with schizophrenia towards the cluster of healthy controls. A striking finding of this study is the effect of the number of psychotic episodes prior to commencing antiphychotic treatment on the CSF metabolite profile in patients with schizophrenia. Of 21 patients who commenced antipsychotic medication during their first psychotic episode, 57% clustered with healthy controls whereas six out of the seven patients who had several psychotic episodes prior to treatment clustered with the group of drug-naïve patients with first-onset schizophrenia. These results suggest that the initiation of antipsychotic treatment during a first psychotic episode may influence treatment response and/or indeed outcome.
Pre-eclampsia is an important cause of maternal morbidity and mortality while the World Health Organization estimates that worldwide over 100,000 women die from pre-eclampsia each year. By using GC-tof-MS the authors [31] were able to separate and detect several hundred metabolites from both control (87) and diseased (87) samples. The application of genetic algorithms on these data indicated that the pre-eclamptic plasma could be discriminated from the matched controls on the basis of just three metabolite peaks (two of which tended to be lower and one tended to be higher in the samples from women with pre-eclampsia, and to a certain extent this correlated with the severity of the disease). In this context it is worth commenting that genetic algorithms is advantageous over other machine-learning methods such as neural networks and support vector machines, as it allows one to understand the problem in terms of small subsets of input variables that it combines into rules. In the case of Kenny and colleagues [31], only 10 of each the disease and control samples were taken at a gestational age of under 30 weeks, and a clear task for the future is to establish the extent to which these diagnostic rules apply earlier in pregnancy and thus are of greater prognostic value.
A metabolic “bioprofile” consisting of predictive serum metabolite features from 1H NMR spectral data of the murine K/BxN model of arthritis were presented in the study of Weljie et al. [32]. A unique method was developed by combining technologies such as quantitative targeted profiling, O-PLS-DA pattern recognition analysis and metabolic-pathway-based network analysis for interpretation of results. In total, 88 spectral features were profiled (59 metabolites and 28 unknown resonances). A highly significant subset of 18 spectral features (15 known compounds and 3 unknown resonances) was identified and in this metabolic bioprofile, metabolites relating to nucleic acid, amino acid, and fatty acid metabolism, as well as lipolysis, reactive oxygen species generation, and methylation were among them. Pathway analysis suggested a shift from metabolites involved in numerous reactions (hub metabolites) toward intermediates and metabolic endpoints associated with arthritis.
4. Metabolomics in Drug Discovery and Polypharmacology Studies
Drug molecules generally act on specific targets at the cellular level, and upon binding to the receptors, they exert a desirable alteration of the cellular activities, regarded as the pharmaceutical effect. Current drug discovery depends largely on ransom screening, either high-throughput screening (HTS) in vitro, or virtual screening (VS) in silico. Because the number of available compounds is huge, several drug-likeness filters are proposed to reduce the number of compounds that need to be evaluated. The ability to effectively predict if a chemical compound is “drug-like” or “nondrug-like” is, thus, a valuable tool in the design, optimization, and selection of drug candidates for development [33]. Drug-likeness is a general descriptor of the potential of a small-molecule to become a drug. It is not a unified descriptor but a global property of a compound processing many specific characteristics such as good solubility, membrane permeability, half-life, and having a pharmacophore pattern to interact specifically with a target protein. These characteristics can be reflected as molecular descriptors such as molecular weight, log P, the number of hydrogen-bond donors, the number of hydrogen-bond acceptors, the number of rotatable bonds, the number of rigid bonds, the number of rings in a molecule, and so forth [34]. Lipinski's widely used rule of 5 defines drug-like “as those compounds that have sufficiently acceptable absorption, distribution, metabolism, excretion, and toxicity (ADMET) properties to survive through the completion of Human Phase I clinical trials” [35]. It has been observed that metabolites tend to obey in their majority the Lipinski “Rule of 5”, which hints to the fact that drugs are indirectly synthesized to mimic the original endogenous substrates [36]. Based on this, metabolite-likeness and biological relevance filters have recently been developed, which consider that chemical compounds from virtual screens of large pharmaceutical libraries that are similar to endogenous metabolites stand more chances for being successful drug candidates [37, 38]. The approach leverages the “chemical similarity principle”, which states that molecules with similar structure likely have similar biological properties.
Drug developers have long-mined small-molecule metabolism for the design of enzyme inhibitors chemically similar to their endogenous substrates. The approach has yielded many successes, including antimetabolites such a folate derivatives used in cancer therapy [39] and the nucleoside analog prodrugs used for antiviral therapy [40]. With the recent availability of databases of metabolites and metabolic reactions, we have gained a wealth of information regarding the interaction of small molecules with biological systems. At the same time, the notion of chemical space and the advance of chemoinformatics tools have paved the way to link the metabolome with structural and physicochemical properties of endogenous metabolites and to predict links between synthetic molecules and human metabolism.
Recent developments in the area of systems biology have lead scientists to realize the limitations of reductionism and begin to lay emphasis on more holistic research patterns, such as systems biology and network pharmacology [41–44], Most diseases are not caused by changes in a single causal gene but by an unbalanced regulating network resulting from the dysfunctions of multiple genes or their products. At the same time, drug molecules commonly participate in biological networks and both their intended effect and side effect are rather systemic than specific to a single biological target.
On this direction, Corey Adams and coworkers have recently demonstrated a new method to predict what enzymes drugs might affect based on the chemical similarity between classes of drugs and the natural chemicals used by enzymes. The authors have applied the method to 246 known drug classes and a collection of 385 organisms to create maps of potential drug action on metabolism. Moreover, they show how the predicted connections can be used to find new ways to kill pathogens and to avoid unintentionally interfering with human enzymes [45].
In the work of Macchiarulo and coworkers, human metabolic pathways are projected and clustered on the chemical space based on similarity of the involved metabolites translated in a set of selected physicochemical and topological descriptors. Further to this, the authors develop a classifier that estimates the proximity of marketed drugs to any given pathway, with the aim to elucidate the extend of overlap and to uncover cross-interactions between drugs and the major human pathways. The model performs well for tightly clustered, isolated pathways, but it loses its predictive ability when it comes to overlapping pathways [46].
5. Metabolomics for the Study of Polypharmacology of Natural Compounds
Internationally, there is a growing and sustained interest from both pharmaceutical companies and public in medicine from natural sources. For the public, natural medicine represent a holistic approach to disease treatment, with potentially less side effects than conventional medicine. For the pharmaceutical companies, bioactive natural products constitute attractive drug leads, as they have been optimized in a long-term natural selection process for optimal interaction with biomolecules. To promote the ecological survival of plants, structures of secondary products have evolved to interact with molecular targets affecting the cells, tissues and physiological functions in competing microorganisms, plants, and animals. In this, respect, some plant secondary products may exert their action by resembling endogenous metabolites, ligands, hormones, signal transduction molecules, or neurotransmitters and thus have beneficial effects on humans due to similarities in their potential target sites [47].
Complementary to the above studies on drug polypharmacology and in order to elucidate the extend of overlap and similarity between natural compounds from plants used in ethnomedicine and human metabolites, we created chemical networks between natural compounds from the Chinese Natural Products Database (CNPD v.2004.1) and human metabolites from HMDB. CNPD is a compilation of 57,346 compounds found in plants largely used in TCM (Traditional Chinese Medicine). These compounds come from 2,611 plant species belonging to 457 different plant genera. After removal of salts, inorganic compounds, and duplicates, we extracted 53,180 unique, organic compounds in SDF format, which we imported into a Molecular Operating Environment (MOE, v.2008.10) [48] database. 1417 of these compounds are annotated with experimentally derived bioactivity information. HMDB v. 2.5 was used as source of human metabolites and 7,985 compounds were extracted in SDF format. All structures were washed, that is all ionizable groups were coordinated with neutral pH conditions, and energy minimized using the MMFF946 force field.
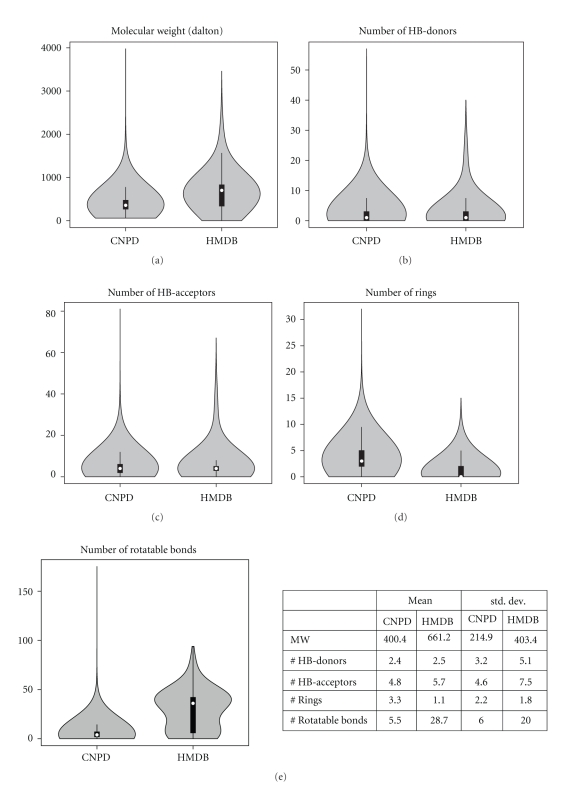
To get a first overview, we compared the two databases considering common descriptors for drug-like molecules, namely molecular weight (MW), number of hydrogen-bond donors (HB donors), number of hydrogen-bond acceptors (HB acceptors), number of rings and number of rotatable bonds. As seen in the violin plots of Figure 3, the human metabolites have higher average molecular weight (MW = 661.2) and broader distribution (std dev = 403.4), which is obviously due to the presence of many lipids (3800 out of 7985 compounds are lipids in the newest version of the HMDB) [49]. The number of HB-donors is almost the same in both CNPD and HMDB sets, with an average value of 2.4 and 2.5, respectively and 90% of the compounds in each data set having ≤5 HB-donors. When it comes to HB-acceptors, the two profiles differ slightly. 90% of the compounds in each data set have ≤10 HB-acceptors, but there is higher percentage of human metabolites with 9-10 HB-acceptors (6.9%) than are natural compounds (4.7%). Due to this, CNPD has a lower mean value and std deviation (4.8 and 4.6, respectively). The number of rings is lower in HMDB, again because of the presence of lipids that are acyclic. As a consequence, compounds from HMDB have on average many more rotatable bonds than their counterparts from CNPD. While 95% of compounds from CNPD have up to 15 rotatable bonds, half of the compounds from HMDB have between 30 and 50 rotatable bonds.
Figure 3.
Comparison of the distribution of selected druglike molecular properties for natural compounds from CNPD and human metabolites from HMDB. Violin plots for (a) molecular weight, (b) hydrogen-bond donors, (c) hydrogen-bond acceptors, (d) number of rings and (e) number of rotatable bonds, along with table with mean values and standard deviations. A violin plot is a combination of a box plot and a kernel density plot and offers a more detailed view of a dataset's variability than a box plot alone. The white marker indicates the median of the data and the black box the interquirtile range (the difference between the third and first quartiles that contain 50% of the distribution). The black lines extend to one and a half times the width of the box. Violin plots were made in R.
Despite the differences noted above, there is a significant room for overlap between the chemical spaces of the two datasets, which we attempt to elucidate via a more thorough structural similarity analysis that follows. First we investigate how many compounds are present in both data sets, by comparing their SMILES (Simplified Molecular Input Line Entry Specification) strings. Following that, we analyze the extend of structural similarity of the two data sets. For this, all pairs of molecules between the two sets are compared using a pairwise similarity metric, which consists of a descriptor and a similarity criterion. For the descriptor, MACCS (Molecular ACCess System) keys were calculated in MOE. The MACCS keys represent each molecule as a vector of 166 bits, each indicating the presence or absence of a predefined substructure or functional group (e.g., aromatic rings, oxygens, amine groups, etc.). The similarity criterion is the widely used Tanimoto coefficient (Tc) [50]. Tc is calculated as shown in (1). If two molecules have a and b bits in their fragment bit-strings, respectively, with c of these bits being present in both their fingerprints, then Tc corresponds to the ratio of the number of bits the two molecules have in common to the total number of occupied bins by both molecules
| (1) |
T c gives valules in the range of zero (no bits in common, 0% similarity) to unity (all bits the same, 100% similarity). The Tc threshold for two compounds being similar was set to 0.85 and the similarity networks were visualized using the Organic Layout of Cytoscape v. 2.6.3 [51].
5.1. Overlap between Human Metabolites and Natural Compounds
There are 383 compounds shared between the two databases, which denotes that, apart from participating in the human metabolism, these natural compounds are secondary metabolites of plants used in ethnopharmacology. For example, 2-pyrocatechuic acid (HMDB00397) is a normal human benzoic acid metabolite found in plasma that is an intermediate of the phenyl propanoid biosynthesis. It is has been isolated from black currant [52], which has long been used in European and Chinese folk medicine as diuretic, treating diarrhea, arthritic pain, and so forth. Recently, 2-pyrocatechuic acid was found to be weak inhibitor of Selectin E [53] and potent inhibitor of 15-lipogygenase-catalysed oxygenation of arachidonic acid that is involved in many aspects of inflammatory disease and in particular in the development of colorectal cancer [54].
Another example, indole (HMDB00738), is an aromatic heterocyclic organic compound that occurs naturally in human feces and has an intense fecal smell. At very low concentrations, however, it has a flowery smell and is a constituent of many flower scents. Natural jasmine oil that contains around 2.5% of indole is used traditionally for healing the female reproducing system, to treat headaches and insomnia. In human metabolism, indole participates in the tryptophan metabolic pathway, which is a highly regulated biological process. There has been significant research on the medical implications involved in dysregulation of tryptophan metabolism. Abnormalities in it may play a role in central nervous system diseases such as acquired immunodeficiency syndrome- (AIDS-) related dementia [55], Huntingtom's disease [56] and psychopathological disorders [57]. In addition, data from the literature suggest that a mechanism dependent on tryptophan catabolism might regulate the immune responses to a number of diseases [58–60].
5.2. Similarity Networks of Human Metabolites and Natural Compounds
There are 15,523 natural compounds in CNPD (29% of the total data set) that have a Tanimoto similarity coefficient of 0.85 or higher with at least one human metabolite. In total, there are formed 233,211 similarity pairs between the two datasets, which indicates that each natural compound is similar—on average—with 15 human metabolites.
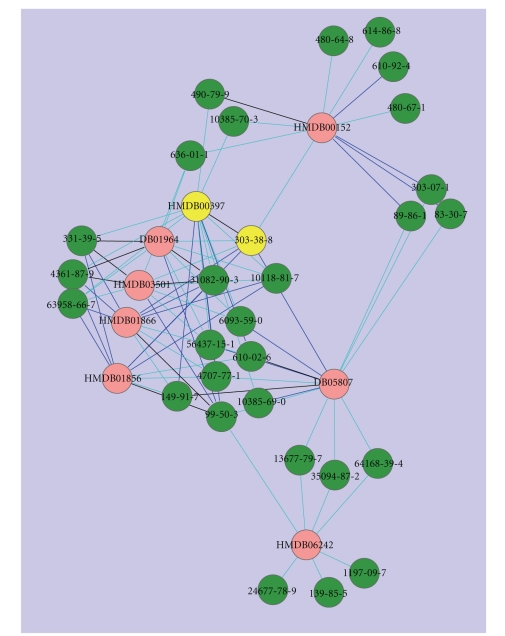
As an illustrative example, Figures 4 and 5 below show the similarity networks of 2-pyrocatechuic acid and indole that were discussed in the previous section. As seen in Figure 4, 2-pyrocatechuic acid is linked with Tc ≥ 0.9 to seven other human metabolites and 28 natural compounds from CNPD. Interestingly, the human metabolites of this similarity network belong to two main metabolic processes. HMDB01866, HMDB06242, HMDA00152, and HMDB01856 are involved in tyrosine metabolism/biosynthesis, while HMDB00397, HMDB03501, and HMDB01964 are intermediates of the phenyl propanoid biosynthesis. Recent research on tyrosine metabolism suggests strong correlation with chronic kidney failure [61], eating disorders and migraine [62]. The natural compounds from CNPD that are met in the network are primarily benzoic acid derivatives from diverse sources of plants (e.g., picea maximowiczii, grevillea robusta), fungi (e.g., polyporus tumulosus, boletus scaber) and flowers (e.g., centaurium erythraea, anthemis nobilis), many of which are known as folk medicine.
Figure 4.
Similarity network of 2-pyrocatechuic acid. Pink nodes indicate human metabolites from HMDB and green nodes indicate natural compounds from CNPD. Node labels denote the respective ID codes of the compounds. The nodes are linked when the two compounds have Tc ≥ 0.85. Due to the high number of pairs with similarity between 0.85 and 0.90, we included in the figure only connections of Tc ≥ 0.90 to allow better visualization of the network. The width and color of the edges are analogous to the value of Tc: Cyan: 0.90 ≤ Tc < 0.95, Blue: 0.95 ≤ Tc < 1.0, Black: Tc = 1. The two nodes in yellow denote 2-pyrocatechuic acid with HMDB ID and CAS registry number, respectively.
Figure 5.
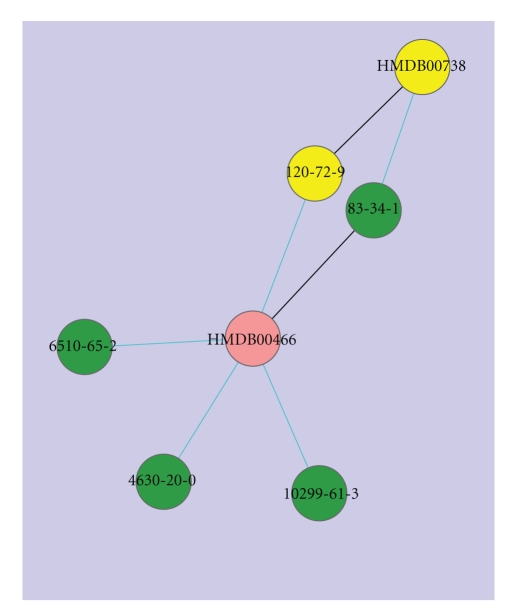
Similarity network of indole. Pink nodes indicate human metabolites from HMDB and green nodes indicate natural compounds from CNPD. Node labels denote the respective ID codes of the compounds. The nodes are linked when the two compounds have Tc ≥ 0.85. The width and color of the edges are analogous to the value of Tc: Cyan: 0.85 ≤ Tc < 0.95, Black: Tc = 1. The node in yellow denotes indole with HMDB ID and CAS registry number, respectively.
Indole, shown in Figure 5, is linked to one other human metabolite, and four natural compounds from CNPD. HMDB00466 is the compound 3-methyl indole that is involved in tryptophan metabolism as well. Three natural compounds from CNPD have high similarity to the two human metabolites. 1-methyl-9H-carbazole (cas: 6510-65-2), 3-methyl-9H-carbazole (cas: 4630-20-0) and 2,4-dimethyl-1H-indole (cas: 10299-61-3) are alkaloids found in Tedania ignis (a sponge species) [63], glycosmis pentaphylla (orangeberry) [64], and Tricholoma virgatum (a muchroom species) [65], respectively. These natural compounds from CNPD that are found similar to well-studied human metabolites are potentially interesting leads with druglike and metabolite-like properties that would be worth investigating further for their medicinal properties and their impact on human health.
In order to evaluate how the 29% similarity of CNPD to HMDB compares with other types of data sets, we performed the same analysis for 4,567 approved and experimental drugs from DrugBank v.2, as well as for a randomly selected subset of 59,025 compounds from ChemDiv, a commercial provider of small compounds for drug discovery HTS. Quite remarkably, the compounds from DrugBank showed the same extend of similarity to HMDB as natural compounds from CNPD. 1,331 drug compounds (29%) were found to be similar to human metabolites, forming 35,635 similarity pairs. On the other hand, only 182 compounds from the subset of ChemDiv were found similar to any human metabolite, forming just 1,563 similarity pairs in total. This low similarity of ChemDiv could be attributed to the fact that HTS databases contain small molecules with simple structures that can be easily modified further to more potent drug candidates. These finding supports the hypothesis that as drug candidates move forward on the drug optimization platform, there is favorable selection towards those that mimic the endogenous substrates. The fact that natural compounds also resemble the latter may indicate that plants with medicinal properties may exert their action via their molecular components resembling human endogenous metabolites.
6. Future Perspectives
Metabolomics, the study of metabolism at the global level, is moving to exciting directions. With the development of more sensitive and advanced instrumentation and computational tools for data interpretation in the physiological context, metabolomics have the potential to impact our understanding of molecular mechanisms of diseases. A state-of-the-art metabolomics study requires knowledge in many areas and especially at the interface of chemistry, biology, and computer science. High-quality samples, improvements in automated metabolite identification, complete coverage of the human metabolome, establishment of spectral databases of metabolites and associated biochemical identities, innovative experimental designs to best address a hypothesis, as well as novel computational tools to handle metabolomics data are critical hurdles that must be overcome to drive the inclusion of metabolomics in all steps of drug discovery and drug development. The examples presented above demonstrated that metabolite profiles reflect both environmental and genetic influences in patients and reveal new links between metabolites and diseases providing needed prognostic, diagnostic, and surrogate biomarkers. The integration of these signatures with other omic technologies is of outmost importance to characterize the entire spectrum of malignant phenotype.
Systems chemical biology networks that assemble and integrate known and predicted links between small compounds of biological relevance, including human metabolites, can have a great potential in pharmaceutical research that could be used in a variety of ways. Novel ligands can be selected on the premise of being similar to endogenous metabolites with the desired bioactivity profile. Pathways for orphan metabolites could be predicted, based on their similarity with compounds of known biological target and mode of action. New ways to kill pathogens and to avoid unintentionally interfering with human enzymes can be investigated and cross-interactions between drugs and the major human pathways can be unravelled. Last but not least, one could predict the biological targets of bioactive natural compounds from medicinal plants, by looking at their similarity networks with human metabolites with known biological targets. By adding information about the metabolic pathways that these metabolites are involved, one could also extract hypotheses regarding the mode of action and therapeutic mechanism of the medicinal plant at the molecular level, which is at the moment the missing link for the coupling of WM with TCM and ethnomedicine in general.
Acknowledgments
The authors acknowledge the funding from the Danish Research Council for Technology and Production Sciences. they thank Sonny Kim Nielsen for the Tanimoto coefficient calculations.
References
- 1.Dettmer K, Hammock BD. Metabolomics—a new exciting field within the ‘omics’ sciences. Environmental Health Perspectives. 2004;112(7):396–397. doi: 10.1289/ehp.112-1241997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schmidt CW. What’s happening downstream of DNA. Environmental Health Perspectives. 2004;112(7):410–415. doi: 10.1289/ehp.112-a410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van der Greef J, Martin S, Juhasz P, et al. The art and practice of systems biology in medicine: mapping patterns of relationships. Journal of Proteome Research. 2007;6(4):1540–1559. doi: 10.1021/pr0606530. [DOI] [PubMed] [Google Scholar]
- 4.Rozen S, Cudkowicz ME, Bogdanov M, et al. Metabolomic analysis and signatures in motor neuron disease. Metabolomics. 2005;1(2):101–108. doi: 10.1007/s11306-005-4810-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paige LA, Mitchell MW, Krishnan KRP, Kaddurah-Daouk R, Steffens DC. A preliminary metabolomic analysis of older adults with and without depression. International Journal of Geriatric Psychiatry. 2007;22(5):418–423. doi: 10.1002/gps.1690. [DOI] [PubMed] [Google Scholar]
- 6.Kaddurah-Daouk R, Kristal BS, Weinshilboum RM. Metabolomics: a global biochemical approach to drug response and disease. Annual Review of Pharmacology and Toxicology. 2008;48:653–683. doi: 10.1146/annurev.pharmtox.48.113006.094715. [DOI] [PubMed] [Google Scholar]
- 7.Holmes E, Tsang TM, Huang JTJ, et al. Metabolic profiling of CSF: evidence that early intervention may impact on disease progression and outcome in schizophrenia. PLoS Medicine. 2006;3(8, article e327) doi: 10.1371/journal.pmed.0030327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaddurah-Daouk R. Metabolic profiling of patients with schizophrenia. PLoS Medicine. 2006;3(8, article e363) doi: 10.1371/journal.pmed.0030363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morvan D, Demidem A. Metabolomics by proton nuclear magnetic resonance spectroscopy of the response to chloroethylnitrosourea reveals drug efficacy and tumor adaptive metabolic pathways. Cancer Research. 2007;67(5):2150–2159. doi: 10.1158/0008-5472.CAN-06-2346. [DOI] [PubMed] [Google Scholar]
- 10.Yang J, Xu G, Zheng Y, et al. Diagnosis of liver cancer using HPLC-based metabonomics avoiding false-positive result from hepatitis and hepatocirrhosis diseases. Journal of Chromatography B. 2004;813(1-2):59–65. doi: 10.1016/j.jchromb.2004.09.032. [DOI] [PubMed] [Google Scholar]
- 11.Fan X, Bai J, Shen P. Diagnosis of breast cancer using HPLC metabonomics fingerprints coupled with computational methods. In: Proceedings of the 27th Annual International Conference of the Engineering in Medicine and Biology Society (EMBS ’05); September 2005; Shanghai, China. pp. 6081–6084. [DOI] [PubMed] [Google Scholar]
- 12.Van Doorn M, Vogels J, Tas A, et al. Evaluation of metabolite profiles as biomarkers for the pharmacological effects of thiazolidinediones in type 2 diabetes mellitus patients and healthy volunteers. British Journal of Clinical Pharmacology. 2007;63(5):562–574. doi: 10.1111/j.1365-2125.2006.02816.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clayton TA, Lindon JC, Cloarec O, et al. Pharmaco-metabonomic phenotyping and personalized drug treatment. Nature. 2006;440(7087):1073–1077. doi: 10.1038/nature04648. [DOI] [PubMed] [Google Scholar]
- 14.Madsen R, Lundstedt T, Trygg J. Chemometrics in metabolomics-a review in human disease diagnosis. Analytica Chimica Acta. 2010;659(1-2):23–33. doi: 10.1016/j.aca.2009.11.042. [DOI] [PubMed] [Google Scholar]
- 15.Kramer R. Chemometric Techniques for Quantitative Analysis. New York, NY, USA: Marcel Dekker; 1998. [Google Scholar]
- 16.Kohonen T. Self-organized formation of topologically correct feature maps. Biological Cybernetics. 1982;43(1):59–69. [Google Scholar]
- 17.Meyer D, Leisch F, Hornik K. The support vector machine under test. Neurocomputing. 2003;55(1-2):169–186. [Google Scholar]
- 18.Müller P, Insua DR. Issues in Bayesian analysis of neural network models. Neural Computation. 1998;10(3):749–770. doi: 10.1162/089976698300017737. [DOI] [PubMed] [Google Scholar]
- 19.Guan W, Zhou M, Hampton CY, et al. Ovarian cancer detection from metabolomic Liquid chromatography/mass spectrometry data by support vector machines. BMC Bioinformatics. 2009;10, article 259 doi: 10.1186/1471-2105-10-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henneges C, Bullinger D, Fux R, et al. Prediction of breast cancer by profiling of urinary RNA metabolites using Support Vector Machine-based feature selection. BMC Cancer. 2009;9, article 104 doi: 10.1186/1471-2407-9-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arakaki AK, Mezencev R, Bowen NJ, Huang Y, McDonald JF, Skolnick J. Identification of metabolites with anticancer properties by computational metabolomics. Molecular Cancer. 2008;7, article 57 doi: 10.1186/1476-4598-7-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Southam AD, Easton JM, Stentiford GD, Ludwig C, Arvanitis TN, Viant MR. Metabolic changes in flatfish hepatic tumours revealed by NMR-based metabolomics and metabolic correlation networks. Journal of Proteome Research. 2008;7(12):5277–5285. doi: 10.1021/pr800353t. [DOI] [PubMed] [Google Scholar]
- 23.Altmaier E, Ramsay SL, Graber A, Mewes H-W, Weinberger KM, Suhre K. Bioinformatics analysis of targeted metabolomics—uncovering old and new tales of diabetic mice under medication. Endocrinology. 2008;149(7):3478–3489. doi: 10.1210/en.2007-1747. [DOI] [PubMed] [Google Scholar]
- 24.Wang C, Kong H, Guan Y, et al. Plasma phospholipid metabolic profiling and biomarkers of type 2 diabetes mellitus based on high-performance Liquid chromatography/electrospray mass spectrometry and multivariate statistical analysis. Analytical Chemistry. 2005;77(13):4108–4116. doi: 10.1021/ac0481001. [DOI] [PubMed] [Google Scholar]
- 25.Mäkinen V-P, Soininen P, Forsblom C, et al. 1H NMR metabonomics approach to the disease continuum of diabetic complications and premature death. Molecular Systems Biology. 2008;4, article 167 doi: 10.1038/msb4100205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salek RM, Maguire ML, Bentley E, et al. A metabolomic comparison of urinary changes in type 2 diabetes in mouse, rat, and human. Physiological Genomics. 2007;29(2):99–108. doi: 10.1152/physiolgenomics.00194.2006. [DOI] [PubMed] [Google Scholar]
- 27.Connor SC, Hansen MK, Corner A, Smith RF, Ryan TE. Integration of metabolomics and transcriptomics data to aid biomarker discovery in type 2 diabetes. Molecular Biosystems. 2010;6:909–921. doi: 10.1039/b914182k. [DOI] [PubMed] [Google Scholar]
- 28.Lanza IR, Zhang S, Ward LE, Karakelides H, Raftery D, Nair KS. Quantitative metabolomics by H-NMR and LC-MS/MS confirms altered metabolic pathways in diabetes. PLoS One. 2010;5:e10538 pages. doi: 10.1371/journal.pone.0010538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bao Y, Zhao T, Wang X, et al. Metabonomic variations in the drug-treated type 2 diabetes mellitus patients and healthy volunteers. Journal of Proteome Research. 2009;8:1623–1630. doi: 10.1021/pr800643w. [DOI] [PubMed] [Google Scholar]
- 30.Čuperlović-Culf M, Belacel N, Culf AS, et al. NMRmetabolic analysis of samples using fuzzy K-means clustering. Magnetic Resonance in Chemistry. 2009;47(supplement 1):96–104. doi: 10.1002/mrc.2502. [DOI] [PubMed] [Google Scholar]
- 31.Kenny LC, Dunn WB, Ellis DI, Myers J, Baker PN, Kell DB. Novel biomarkers for pre-eclampsia detected using metabolomics and machine learning. Metabolomics. 2005;1(3):227–234. [Google Scholar]
- 32.Weljie AM, Dowlatabadi R, Miller BJ, Vogel HJ, Jirik FR. An inflammatory arthritis-associated metabolite biomarker pattern revealed by 1H NMR spectroscopy. Journal of Proteome Research. 2007;6(9):3456–3464. doi: 10.1021/pr070123j. [DOI] [PubMed] [Google Scholar]
- 33.Zhang M-Q, Wilkinson B. Drug discovery beyond the ‘rule-of-five’. Current Opinion in Biotechnology. 2007;18(6):478–488. doi: 10.1016/j.copbio.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 34.Willett P. Similarity-based virtual screening using 2D fingerprints. Drug Discovery Today. 2006;11(23-24):1046–1053. doi: 10.1016/j.drudis.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 35.Lipinski CA. Drug-like properties and the causes of poor solubility and poor permeability. Journal of Pharmacological and Toxicological Methods. 2000;44(1):235–249. doi: 10.1016/s1056-8719(00)00107-6. [DOI] [PubMed] [Google Scholar]
- 36.Nobeli I, Ponstingl H, Krissinel EB, Thornton JM. A structure-based anatomy of the E. coli metabolome. Journal of Molecular Biology. 2003;334(4):697–719. doi: 10.1016/j.jmb.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 37.Gupta S, Aires-de-Sousa J. Comparing the chemical spaces of metabolites and available chemicals: models of metabolite-likeness. Molecular Diversity. 2007;11(1):23–36. doi: 10.1007/s11030-006-9054-0. [DOI] [PubMed] [Google Scholar]
- 38.Kong D-X, Ren W, Lu W, Zhang H-Y. Do biologically relevant compounds have more chance to be drugs? Journal of Chemical Information and Modeling. 2009;49(10):2376–2381. doi: 10.1021/ci900229c. [DOI] [PubMed] [Google Scholar]
- 39.Scagliotti GV, Selvaggi G. Antimetabolites and cancer: emerging data with a focus on antifolates. Expert Opinion on Therapeutic Patents. 2006;16(2):189–200. doi: 10.1517/13543776.16.2.189. [DOI] [PubMed] [Google Scholar]
- 40.Meerbach A, Meier C, Sauerbrei A, Meckel H-M, Wutzler P. Antiviral activity of cyclosaligenyl prodrugs of the nucleoside analogue bromovinyldeoxyuridine against herpes viruses. International Journal of Antimicrobial Agents. 2006;27(5):423–430. doi: 10.1016/j.ijantimicag.2005.11.021. [DOI] [PubMed] [Google Scholar]
- 41.Oprea TI, Tropsha A, Faulon J-L, Rintoul MD. Systems chemical biology. Nature Chemical Biology. 2007;3(8):447–450. doi: 10.1038/nchembio0807-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Berger SI, Iyengar R. Network analyses in systems pharmacology. Bioinformatics. 2009;25(19):2466–2472. doi: 10.1093/bioinformatics/btp465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schadt EE, Friend SH, Shaywitz DA. A network view of disease and compound screening. Nature Reviews Drug Discovery. 2009;8(4):286–295. doi: 10.1038/nrd2826. [DOI] [PubMed] [Google Scholar]
- 44.Zhao J, Jiang P, Zhang W. Molecular networks for the study of TCM pharmacology. Briefings in Bioinformatics. 2010;11(4):417–430. doi: 10.1093/bib/bbp063. [DOI] [PubMed] [Google Scholar]
- 45.Adams JC, Keiser MJ, Basuino L, et al. A mapping of drug space from the viewpoint of small molecule metabolism. PLoS Computational Biology. 2009;5(8) doi: 10.1371/journal.pcbi.1000474. Article ID e1000474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Macchiarulo A, Thornton JM, Nobeli I. Mapping human metabolic pathways in the small molecule chemical space. Journal of Chemical Information and Modeling. 2009;49(10):2272–2289. doi: 10.1021/ci900196u. [DOI] [PubMed] [Google Scholar]
- 47.Briskin DP. Medicinal plants and phytomedicines. Linking plant biochemistry and physiology to human health. Plant Physiology. 2000;124(2):507–514. doi: 10.1104/pp.124.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chemical Computing Group. Molecular Operating Environment. Montreal, Canada: 2008. [Google Scholar]
- 49.Wishart DS, Knox C, Guo AC, et al. HMDB: a knowledgebase for the human metabolome. Nucleic Acids Research. 2009;37(1):603–610. doi: 10.1093/nar/gkn810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tanimoto T. IBM internal report. 1957.
- 51.Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software Environment for integrated models of biomolecular interaction networks. Genome Research. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zadernowski R, Naczk M, Nesterowicz J. Phenolic acid profiles in some small berries. Journal of Agricultural and Food Chemistry. 2005;53(6):2118–2124. doi: 10.1021/jf040411p. [DOI] [PubMed] [Google Scholar]
- 53.Kranich R, Busemann AS, Bock D, et al. Rational design of novel, potent small molecule pan-selectin antagonists. Journal of Medicinal Chemistry. 2007;50(6):1101–1115. doi: 10.1021/jm060536g. [DOI] [PubMed] [Google Scholar]
- 54.Russell WR, Scobbie L, Duthie GG, Chesson A. Inhibition of 15-lipoxygenase-catalysed oxygenation of arachidonic acid by substituted benzoic acids. Bioorganic and Medicinal Chemistry. 2008;16(8):4589–4593. doi: 10.1016/j.bmc.2008.02.041. [DOI] [PubMed] [Google Scholar]
- 55.Ruddick JP, Evans AK, Nutt DJ, Lightman SL, Rook GAW, Lowry CA. Tryptophan metabolism in the central nervous system: medical implications. Expert Reviews in Molecular Medicine. 2006;8(20):1–27. doi: 10.1017/S1462399406000068. [DOI] [PubMed] [Google Scholar]
- 56.Stoy N, Mackay GM, Forrest CM, et al. Tryptophan metabolism and oxidative stress in patients with Huntington’s disease. Journal of Neurochemistry. 2005;93(3):611–623. doi: 10.1111/j.1471-4159.2005.03070.x. [DOI] [PubMed] [Google Scholar]
- 57.Zignego AL, Cozzi A, Carpenedo R, et al. HCV patients, psychopathology and tryptophan metabolism: analysis of the effects of pegylated interferon plus ribavirin treatment. Digestive and Liver Disease. 2007;39(1):107–111. doi: 10.1016/s1590-8658(07)80021-1. [DOI] [PubMed] [Google Scholar]
- 58.Torres MI, Lopez-Casado MA, Lorite P, Rios A. Tryptophan metabolism and indoleamine 2,3-dioxygenase expression in coeliac disease. Clinical and Experimental Immunology. 2007;148(3):419–424. doi: 10.1111/j.1365-2249.2007.03365.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Meyer KC, Arend RA, Kalayoglu MV, Rosenthal NS, Byrne GI, Brown RR. Tryptophan metabolism in chronic inflammatory lung disease. Journal of Laboratory and Clinical Medicine. 1995;126(6):530–540. [PubMed] [Google Scholar]
- 60.Schrocksnadel K, Wirleitner B, Winkler C, Fuchs D. Monitoring tryptophan metabolism in chronic immune activation. Clinica Chimica Acta. 2006;364(1-2):82–90. doi: 10.1016/j.cca.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 61.Kopple JD. Phenylalanine and tyrosine metabolism in chronic kidney failure. Journal of Nutrition. 2007;137(6):1586–1598. doi: 10.1093/jn/137.6.1586S. [DOI] [PubMed] [Google Scholar]
- 62.D’Andrea G, Ostuzzi R, Bolner A, et al. Study of tyrosine metabolism in eating disorders. Possible correlation with migraine. Journal of the Neurological Sciences. 2008;29(1):88–92. doi: 10.1007/s10072-008-0895-4. [DOI] [PubMed] [Google Scholar]
- 63.Dillman RL, Cardellina JH., II Aromatic secondary metabolites from the sponge Tedania ignis. Journal of Natural Products. 1991;54(4):1056–1061. [Google Scholar]
- 64.Garlaschelli L, Pang Z, Sterner O, Vidari G. New indole derivatives from the fruit bodies of Tricholoma sciodes and T. virgatum. Tetrahedron. 1994;50(11):3571–3574. [Google Scholar]
- 65.Chowdhury BK, Mustapha A, Garba M, Bhattacharyya P. Carbazole and 3-methylcarbazole from Glycosmis pentaphylla. Phytochemistry. 1987;26(7):2138–2139. [Google Scholar]