Figure 1.
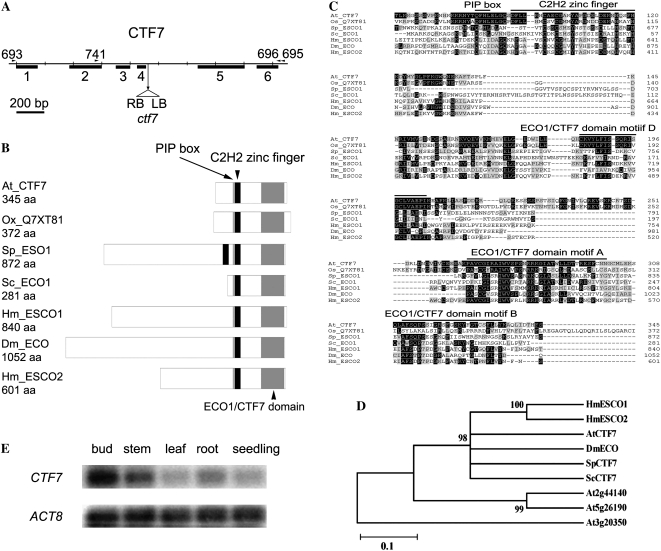
Arabidopsis CTF7 locus and protein structure. A, Gene map of CTF7. The exon positions and T-DNA insertion site are shown. Primers used in this study are shown above the map. LB, Left border; RB, right border. B, Schematic representations of ECO1/CTF7 proteins from different organisms. The ECO1/CTF7 domain (gray box), C2H2 zinc finger domain (black box), and PIP box (thin gray line) are shown. Accession numbers are as follows: Arabidopsis CTF7 (EU077499), O. sativa CTF7 (Q7XY81), S. pombe Eso1 (O42917), S. cerevisiae Eco1 (P43605), Homo sapiens Esco1 (Q5FWF5), Drosophila Eco (Q9VS50), H. sapiens Esco2 (Q56NI9). aa, Amino acids. C, Alignment of ECO1/CTF7 proteins in different organisms. The conserved PIP box, C2H2 zinc finger motif, and acetyltransferase domain are overlined. Identical and similar amino acids are shaded black and gray, respectively. D, Phylogenetic tree of characterized ECO1/CTF7 proteins. E, CTF7 transcripts in different tissues of Arabidopsis. ACTIN8 (ACT8) transcripts were used as a control.