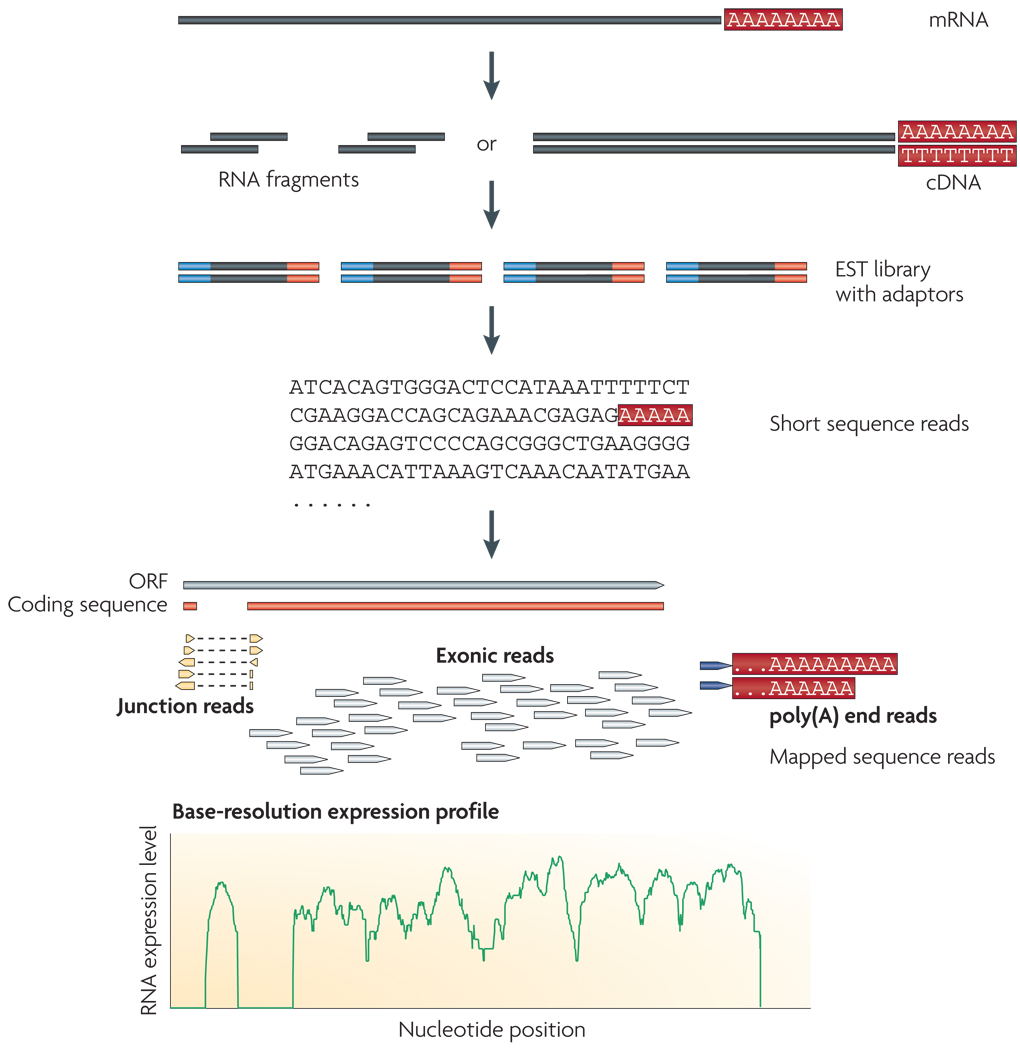
Figure 1. A typical RNA-Seq experiment.
Briefly, long RNAs are first converted into a library of cDNA fragments through either RNA fragmentation or DNA fragmentation (see main text). Sequencing adaptors (blue) are subsequently added to each cDNA fragment and a short sequence is obtained from each cDNA using high-throughput sequencing technology. The resulting sequence reads are aligned with the reference genome or transcriptome, and classified as three types: exonic reads, junction reads and poly(A) end-reads. These three types are used to generate a base-resolution expression profile for each gene, as illustrated at the bottom; a yeast ORF with one intron is shown.