SUMMARY
The HIV-1 Virion infectivity factor (Vif) inhibits the innate viral immunity afforded by the APOBEC3 family of cytidine deaminases. Vif targets the APOBEC3 family for poly-ubiquitination and subsequent proteasomal degradation by linking the Elongin BC dependent ubiquitin ligase complex with the APOBEC3 proteins. The interaction between Vif and the heterodimeric Elongin BC complex, which is mediated by Vif’s viral SOCS (suppressor of cytokine signaling) box, is essential for Vif function. The biophysical consequences of the full length Vif:Elongin BC interaction have not been extensively reported. In this study hydrogen exchange mass spectrometry (HX MS) was used to dissect the Vif:Elongin BC interaction. Elongin C was found to be highly dynamic in the Elongin BC complex while Elongin B was much more stable. Recombinant full length Vif interacted with the Elongin BC complex in vitro with a Kd of 1.9 μM and resulted in observable changes in deuterium uptake in both Elongin C and B. Upon binding to Elongin BC, no significant global conformational changes were detected in Vif by HX MS, but a short fragment of Vif that consisted of the viral SOCS box showed decreased deuterium incorporation upon Elongin BC incubation, suggesting that this region folds upon binding.
Keywords: Hydrogen exchange mass spectrometry, protein conformation, Viral SOCS box, APOBEC3, E3 ubiquitin ligase
The Elongin BC complex is an essential component of the Elongin BC dependent E3 ubiquitin ligase complex. Human Elongin B and C were originally identified as part of the heterotrimeric transcription factor Elongin ABC complex 1 which promotes elongation during transcription by suppressing pausing of RNA polymerase II.2 Elongin B and C also have the ability to form the heterodimeric Elongin BC complex which associates with cellular proteins that contain a Suppressor Of Cytokine Signaling (SOCS) box motif.3–5 Upon binding SOCS-containing proteins (such as SOCS1-7, CIS), the Elongin BC complex contributes to modulation of intracellular responses from various cytokines by bridging SOCS family of proteins and their specific cytokine related targets with machinery of the E3 ubiquitin ligase for proteasomal targeting and subsequent degradation. 5 HIV-1 Vif exploits the Elongin BC dependent E3 ubiquitin ligase complex for the targeted degradation of specific cellular antiviral factors such as APOBEC3G.6–8
Vif is an accessory HIV gene product that is essential for viral replication.9 Vif blocks the innate antiviral activity of cytidine deaminases of the APOBEC3 family by directing them to proteasomal degradation through an Elongin BC dependent E3 ubiquitin ligase.8; 10–13 Mutations in Vif that prevent association with the Elongin BC complex disrupt proteasomal targeting and degradation of the APOBEC3 proteins (such as A3G, A3F) resulting in a non-productive HIV infection.6; 8; 14 The importance of the Elongin BC dependent Vif ubiquitin ligase complex for destruction of innate cellular immunity against HIV makes understanding the Vif:Elongin BC complex, both structurally and biophysically, extremely important.
To date it has been difficult to characterize the Vif:Elongin BC complex due to difficulties in producing soluble full length Vif and other components of the E3 ligase complex in the relatively high concentrations required for traditional biophysical analysis. In addition, conformational changes in Elongin C when it is part of the Elongin BC complex have not been extensively studied, likely due to the inability of cellular SOCS box sequences (SOCS 1-7) to associate with the Elongin BC complex in vitro15. The existing biophysical data for Vif and Elongin BC are, nonetheless, enlightening. The crystal structure of Elongin BC with a fragment (residues 140–156) of Vif (HXB3) 12 provides insight into how the novel SOCS box in Vif associates with Elongin C in the Elongin BC complex, but does not reveal conformational dynamics and biophysical consequences of this interaction nor report on conformational properties of the remainder of Vif in the complex. Recently Reingewertz et al prepared a synthetic version of the C-terminal domain of Vif and found experimentally that it was unfolded in the absence of interacting proteins like the Elongin BC complex 16 and unfolded based on computation.17 Upon incubation with an Elongin C peptide, no structural changes in the C terminal domain of Vif were detected.16 Elongin C, on the other hand, underwent significant conformational changes when it was incubated (in the absence of Elongin B) with either sequences containing a SOCS box motif (such as VHL) or Elongin A.18
In the present study, hydrogen exchange (HX) monitored by mass spectrometry (MS) was used to investigate the conformational dynamics of the Elongin BC complex and the interaction with Vif in vitro. HX MS is a biophysical technique that is amenable to studying proteins and protein complexes where only small quantities of material are available.19 HX MS therefore, seemed highly suited to the study of Vif and Elongin because it does not require large amounts of protein at high concentration but is still very sensitive to conformational changes as a result of protein-protein interactions.20–21 Using HX MS, we determined that Elongin C was dynamic in solution even when associated with Elongin B. On the other hand, relative to Elongin C, Elongin B was conformationally stable in the Elongin BC complex and only the C-terminal tail was highly dynamic. In experiments where Elongin BC bound to recombinant full length HIV-1 Vif, there were significant changes to deuterium incorporation in Elongin C localized to the Vif binding interface. The stoichiometry of the wt Vif :Elongin BC interaction was found to be 1:1, of moderate affinity, and required no additional cellular components for assembly in vitro. The Vif:Elongin BC interaction was specific and required the C-terminal viral BC box of Vif.
RESULTS AND DISCUSSION
HIV-1 Vif interacts with the Elongin BC complex and alters Elongin C conformation
Although members of the human cellular SOCS protein family (SOCS1-7, CIS) bind to the Elongin BC complex when co-expressed in vivo, cellular SOCS box proteins have been reported to not bind Elongin BC in vitro.15 HIV-1 Vif is an exception, as the affinity of short fragments of Vif and the Elongin BC complex in vitro were recently reported by two different groups 22–23, the affinity of full-length HIV-1 Vif for the Elongin BC complex has not been reported. To investigate whether full length recombinant HIV-1 Vif could bind Elongin BC in vitro we utilized HX MS.
Hydrogen exchange at backbone amide hydrogen positions is sensitive to protein complex formation in that exchange rates are generally slower in proteins that are bound to one another.24 The hydrogens at backbone amide positions in proteins are in continuous flux with hydrogens in the solvent. By replacing ordinary aqueous solvent containing hydrogen with aqueous solvent containing 99.9 mole% deuterium (2H, an isotope of hydrogen), HX can be followed because deuterium has different spectral characteristics than hydrogen. When assessing HX by MS, one monitors changes in mass with increasing time in deuterium.25 Solvent shielding and changes in hydrogen bonding are the two main factors that can change backbone amide HX rates in proteins upon complexation. When a protein complex forms, the interface between the binding partners is likely to exclude solvent and may reduce the rate of exchange in the binding interface by limiting access to exchange sites in one or more proteins in the complex. Changes in the conformational dynamics of the protein, meaning protein movements in solution such as protein breathing, localized unfolding, etc., may also occur upon complexation and can alter the hydrogen bonding network or exposure to solvent such that HX rates are changed. Changes in protein dynamics may occur at the binding interface or elsewhere as a result of structural changes that can be communicated through the protein molecule (e.g., Refs 26–27). Complex formation may be probed with MS simply by measuring the amount of deuterium incorporated into members of a protein complex when alone and comparing the results to deuterium levels for the same protein members when part of a complex.
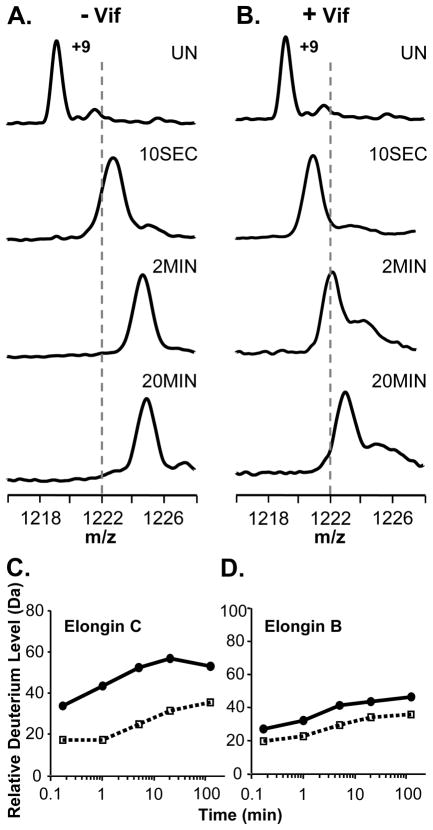
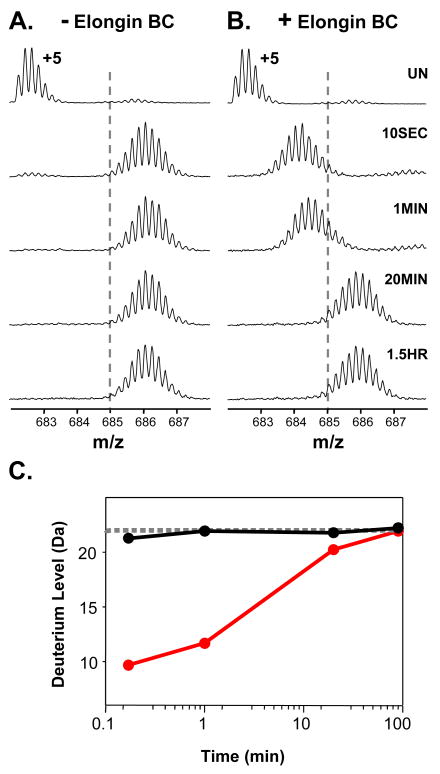
The ability of full-length recombinant Vif to associate with the Elongin BC complex was assayed with HX MS by incubating Elongin BC with a four-fold molar excess of full-length Vif and monitoring the deuterium incorporation in both Elongin B and C. Figure 1 shows the +9 charge state of Elongin C in the Elongin BC complex as it became deuterated with or without Vif. In the presence of Vif, the mass increase of Elongin C was suppressed as a result of complexation. The deuterium uptake curves for Elongin C (Figure 1C) show that a large portion of Elongin C, nearly 20 residues, was protected from hydrogen exchange upon complexation with Vif. Several factors could contribute to such protection including stabilization of the Elongin C structure or the protection of backbone amide hydrogens in Elongin C by the presence of Vif. In addition to monitoring Elongin C, the conformational affects of Vif on Elongin B were also probed. Deuterium incorporation into Elongin B (Figure 1D) was not as dramatically affected by Vif as was Elongin C but Elongin B did show a slight decrease in deuterium uptake upon incubation with Vif. This decrease in deuterium content of Elongin B could propagate from the conformational changes in Elongin C, perhaps as a result of changes at the Elongin BC heterodimeric interface. Taken together these results indicate that upon incubation with full length recombinant HIV-1 Vif in vitro, the conformation and/or dynamics of both Elongins in the Elongin BC complex is affected.
Figure 1.
Global HX MS analysis of Elongin B and C upon binding HIV-1 Vif. (A) Mass spectra of the +9 charge state of Elongin C in the Elongin BC complex or (B) of Elongin C in the Elongin BC complex incubated with a four-fold molar excess of full-length recombinant HIV-1 Vif. The amount of time the complex was incubated in deuterium is indicated (UN, undeuterated). The dotted lines are placed at an arbitrary m/z value as a visual reference. Relative deuterium uptake curves for (C) Elongin C and (D) Elongin B in the absence (solid line) and presence (dotted line) of four-fold molar excess of full length recombinant Vif. The error of intact HX MS measurements was ± 2 Da. Back exchange has not been corrected for in these experiments (see Material and Methods).
The HIV-1 Vif:Elongin interaction is specific and requires the C-terminal portion of Vif
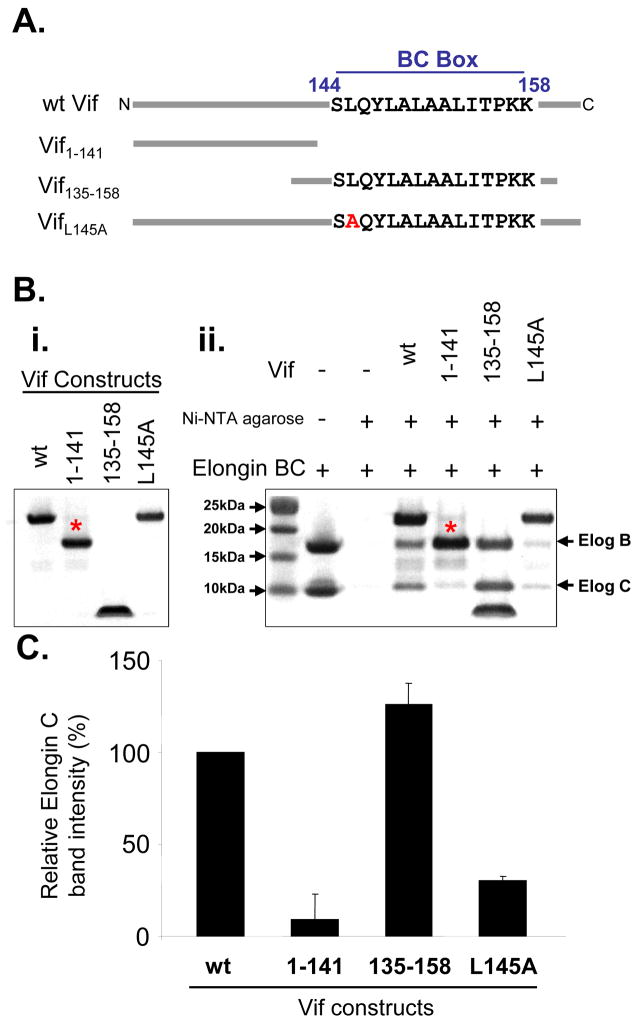
Having established that Vif binds to Elongin BC complex in vitro, we next tested the specificity of the interaction. Several different Vif constructs were analyzed (Figure 2A): wt Vif, Vif1–141, Vif135–158, and VifL145A. These proteins were over-expressed and purified, and their interactions with recombinant Elongin BC complex tested in pull-down experiments (Figure 2B,C) and later with HX MS (see below). Vif1–141 is missing the viral BC box (residues 144–158) which has been shown to be essential for Vif interaction 6. As expected, there was a dramatic reduction in Vif1–141 binding to Elongin BC relative to wt Vif binding (Figure 2B,C). The L145A mutation in Vif was previously shown to reduce viral infectivity 8; 12 and in this in vitro pull-down assay there was a threefold reduction in Elongin BC binding to VifL145A versus wt Vif. We also tested VifΔSLQ (144SLQ146 to AAA) and it showed a nearly identical result to VifL145A (data not shown). Interestingly, Vif135–158 had the most binding to the Elongin BC complex in this assay. This region of Vif contains the residues shown by the Vif140–156: Elongin BC crystal structure 12 to form the first helix of the viral SOCS box. The Vif140–156 crystal structure does not contain any electron density for residues down-stream of the first helix. In other SOCS proteins (e.g., SOCS-2 and 4) this additional C-terminal sequence forms two more helices.3; 5 Therefore the remaining residues of the Vif SOCS box C-terminal to the Viral BC box could interact with the Elongin BC complex via contacts not apparent in the crystal structure and explain the partial binding observed when residues 144SLQ146 are mutated in full length Vif. Our results are in agreement with a recent report showing that Vif139–192 L145A was still able to interact with the Elongin BC complex.22 Taken together, our results indicate that deletion (Vif1–141) and not mutation (VifL145A) of one or several residues in the viral BC box of Vif abolishes the interaction with Elongin BC in vitro and that recombinant Vif and Elongin BC are functionally active and able to interact with one another.
Figure 2.
Parts of recombinant Vif can associate with Elongin BC. (A) Vif constructs used for testing the in vitro interaction with the Elongin BC complex (illustration not to scale). (B) Commassie stained SDS-PAGE gels of (i) the recombinant Vif constructs (1 nmol loaded per lane) and the (ii) pull down analysis between Vif constructs and the Elongin BC complex (3 nmols of Elongin BC were used in each pull-down). Binding was assessed using the Elongin C band due to interference between Elongin B and Vif1–141 bands running at the same apparent molecular weight (as indicated by the *). (C) Densitometry measurements of the Elongin C band in panel B,ii. Relative intensity was normalized to the Elongin C band intensity in the presence of wt Vif. Error bars were determined using two replicate analyses.
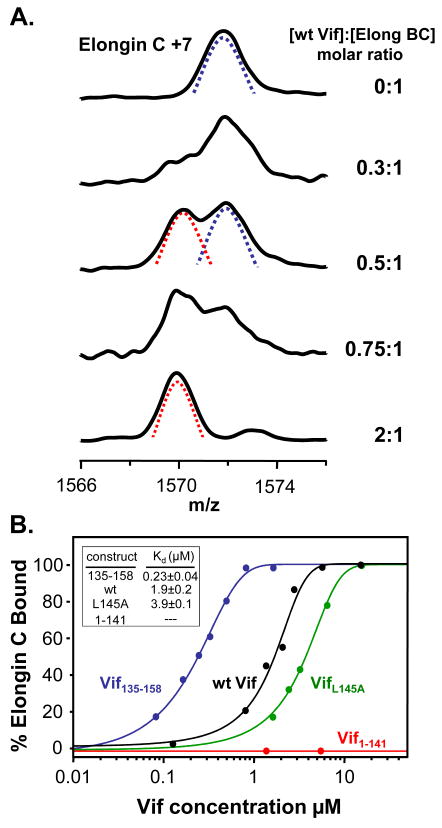
Having established that the Vif:Elongin BC in vitro interaction is specific with recombinant proteins, we used HX MS to determine the dissociation constants between the Elongin BC complex and the different Vif constructs. HX MS can probe dissociation constants for protein-protein and protein-ligand interactions that are not easily assayed by other biophysical techniques.24 The affinity of full length HIV-1 Vif with the components of the E3 ubiquitin ligase has not, to our knowledge, been previously reported. The concentration of the Elongin BC complex was held constant while the concentration of wt Vif was varied. Each concentration combination was labeled with deuterium and the mass of all the proteins measured (as previously described 24). As shown in Figure 3A, it was apparent that as the concentration of Vif increased, more and more protection was afforded to Elongin C. Two populations were apparent in the HX MS data: one that was bound and heavily protected from labeling (Figure 3A, red dotted line) and one that was unbound and not as protected (Figure 3A, blue dotted line). With increasing amounts of Vif, the amount of Elongin C in the unbound state (unprotected and heavily deuterated) decreased and the amount of bound Elongin C (protected and less deuterated) increased. The percentage of bound Elongin C was graphed vs. Vif concentration (μM) and is shown in Figure 3B. Consistent with previous observations,3 the interaction between Elongin B and C in the Elongin BC complex is very strong (it survives cell lysis and both anion exchange and gel filtration chromatography, see Supplementary Material Figure S3); therefore, no dissociation between these two proteins was included in the analysis. For the wt Vif:Elongin BC interaction, an equal molar amount of Vif to Elongin BC was required for 100% bound Elongin C suggesting that the stoichiometry of the wt full-length Vif :Elongin BC interaction was 1:1. The assessment of the binding stoichiometry between HIV-1 Vif and the Elongin BC complex was conducted by fitting the HX MS titration data with a single site binding model. We note that recently, Bergeron et al 23 showed that the C-terminal tail of Elongin B interacted with Vif and therefore the entire Vif:Elongin BC interaction might not be constituted by a single binding site but rather by several regions that involve all three proteins. The HX MS derived dissociation constant for the wt Vif:Elongin BC interaction from duplicate HX MS titration experiments was 1.9 ±0.2 μM. This value is in agreement with a recent ITC report 23 of 1.19 μM for Vif130–180 whereas it is quite different from that in another report 22 where ITC measurements determined the Kd for Vif139–192 to be 0.4 nM. We are unable to explain the large disparity between two of the reports and the third. Although previous studies suggest that VifL145A abolishes the interaction between recombinant Vif and the Elongin BC complex,8; 12 our results indicate that VifL145A still has the ability to interact with the Elongin BC complex in vitro, albeit, with a decreased affinity as compared to wt Vif. VifΔSLQ had a similar affinity as VifL145A (data not shown). Our HX MS derived dissociation constant for VifL145A (3.9 μM ) is in agreement with that recently reported (4.6 μM) for Vif139–192 L145A.22 No binding was detected between Vif1–141 and the Elongin BC complex using our HX MS titration assay, as expected, since Vif1–141 lacks the Viral BC box required for interaction with Elongin BC. Vif135–158 showed the highest affinity for the Elongin BC complex (0.23 μM), nearly a 10-fold increase in affinity for the Elongin BC complex as compared to wt Vif. The trends observed with HX MS titration were consistent with our pull-down analyses (Figure 2) and it is clear that recombinant full-length Vif has the ability to interact with Elongin BC in vitro. The additional residues in full-length Vif somehow contribute to decreased Vif binding to the BC complex, perhaps by conformationally restricting the ability of the disordered C-terminal portion of Vif (in the absence of Cullin 5 and Elongin BC16) to adopt structure capable of binding. The shortened version, Vif135–158, with its higher affinity may more easily adopt a conformation competent with binding than the full-length form or perhaps other portions of the full-length form interfere with binding. Regardless of the differences in apparent affinities between Vif proteins and the Elongin BC complex, the ability of Vif to bind the Elongin BC complex in vitro, whereas other cellular SOCS box containing proteins require co-expression with Elongin BC, might be a unique property of Vif allowing Vif to escape an additional regulatory mechanism that prevents cellular SOCS proteins from binding with Elongin BC without the aid of other cellular components. This property of Vif ensures that HIV-1 has the ability to hijack the E3 ligase machinery to maintain effective viral replication.
Figure 3.
. Vif:Elongin titration. (A) Representative HX MS spectra of the +7 charge state of Elongin C in the presence of wt Vif at various concentration ratios, as indicated on the right. Mass spectra for the other Vif construct titrations can be found in Figure S2. The concentration of Elongin C was fixed at 3.22 μM. The species representing the unbound and therefore more deuterated state of Elongin C is indicated with the blue dotted line. The species representing the bound and therefore protected from deuteration state of Elongin C is indicated with the red dotted line. (B) Titration curves derived from HX MS spectra and the resulting dissociation constants for each construct. The VifΔSLQ mutant had an identical curve to that of VifL145A (data not shown). Vif1–141 (red curve), which lacks the viral BC box, did not bind to the Elongin BC complex in vitro as assayed using HX MS. Each data-point was derived from an HX MS experiment; the lines represent a simple sigmoidal function fit to the data-points. These data are all consistent with the pull-down results in Figure 2.
Localizing conformational flexibility in the Elongin BC complex
The Elongin C structure in solution has been shown to be dynamic and unstable.18; 28–29 The relevance of these findings in the context of the Elongin BC bound to Vif have not been previously reported probably as a result of the high conformational flexibility of Elongin C, the dynamic C-terminal domain of Elongin B, and difficulty in producing suitable quantities of soluble Vif. We utilized HX MS with pepsin digestion (reviewed in 20) to localize which regions of Elongin B and Elongin C were dynamic and determine how these regions may change conformational flexibility in the presence of Vif. Simply, pepsin is used to cleave the deuterium labeled protein into fragments and the deuterium incorporation in each of the fragments is measured as a function of labeling time. Because the sequence of each fragment can be determined, deuterium incorporation can be localized to each peptic peptide. Note that all deuterium exchange is quenched prior to digestion so the amount of deuterium in each peptide reports on the conformation of the protein under physiological labeling conditions.
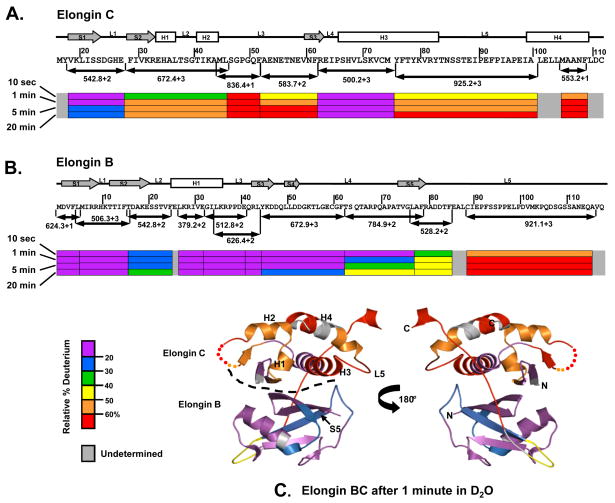
Figure 4 summarizes the results of the pepsin digestion experiments. The regions where Elongin C makes contacts with Elongin B in the formation of the Elongin BC heterodimer showed a slow incorporation of deuterium, consistent with a high degree of solvent protection or conformation stability. These regions in Elongin C included the first beta sheet (S1), loop one (L1) and a section of helix three (H3) which were each ≤ 30% deuterated after 20 minutes in 2H2O. Residues 29–62 in the N-terminal region of Elongin C, including S2, S3, H1, H2, L2 and L3, were ≥ 50% deuterated after 20 minutes in 2H2O. The C-terminal ligand binding domain of Elongin C, including a section H3, L5 and H4, was also heavily deuterated. In contrast, most of Elongin B was quite protected from deuteration implying much more conformational stability. Residues 1–85 of Elongin B are part of the N-terminal ubiquitin-like domain of Elongin B, while residues 86–118 constitute the C-terminal tail.30 As shown in Figure 4B, residues 1–62 were ≤40% deuterated after 20 minutes in 2H2O. The C-terminal portion of Elongin B, however, was much more easily deuterated, i.e., residues 88–116 become ≥60% deuterated after 20 minutes in 2H2O. These results indicate that the C-terminal portion of Elongin B is quite dynamic, solvent exposed and not hydrogen bonded in solution, and are consistent with the lack of structural data on the C-terminal portion of Elongin B as such a dynamic region would be generally difficult to observe with crystallography.
Figure 4.
Summary of peptide-level HX MS analysis of the Elongin BC complex. Representative peptides spanning the length of the (A) Elongin C and (B) Elongin B proteins are shown as color-coded bars below the sequence. Secondary structural elements are represented above the sequence: boxes for helices (H), arrows for beta sheets (S) and lines for loops (L). The relative deuterium level found in each peptide is indicated with the color code at the four labeling times shown at the far left. (C) Deuterium levels in Elongin BC, after one of minute of hydrogen exchange, mapped onto the X-ray structure of Elongin BC (PDB:1LQB).31 The dotted line indicates the boundary between Elongin C and Elongin B. An animation showing deuterium incorporation at all exchange points can be found in the Supplementary Material, Movie S1.
Mapping of the HX MS data for the Elongin BC complex onto the Elongin BC X-ray structure (PDB:1LQB )31 is shown in Figure 4C (see also Supplementary Material Movie S1). The high degree of deuteration in the Elongin C protein when part of the Elongin BC complex suggests that Elongin C is highly flexible in solution and may not be as well ordered as it appears in the crystal. Numerous 3D structures of Elongin BC bound to SOCS box containing proteins have been reported (e.g. 3–4; 12) but to date there is no published structure of the Elongin BC heterodimer without a SOCS box protein bound. We suggest that binding to a SOCS box protein helps to “lock down” Elongin C and stabilize it such that crystallization and high-quality diffraction can occur; the conformational flexibility of Elongin C in the unbound state, as observed here, would be enough to prevent crystallization. Although there is evidence in the literature32–34 that recombinant full-length Vif may exist in a multimeric state, the full-length Vif we have prepared and used for these experiments is still capable of binding to and eliciting an effect on Elongin BC that is completely consistent with the structure of Elongin BC.
Vif stabilizes Elongin BC
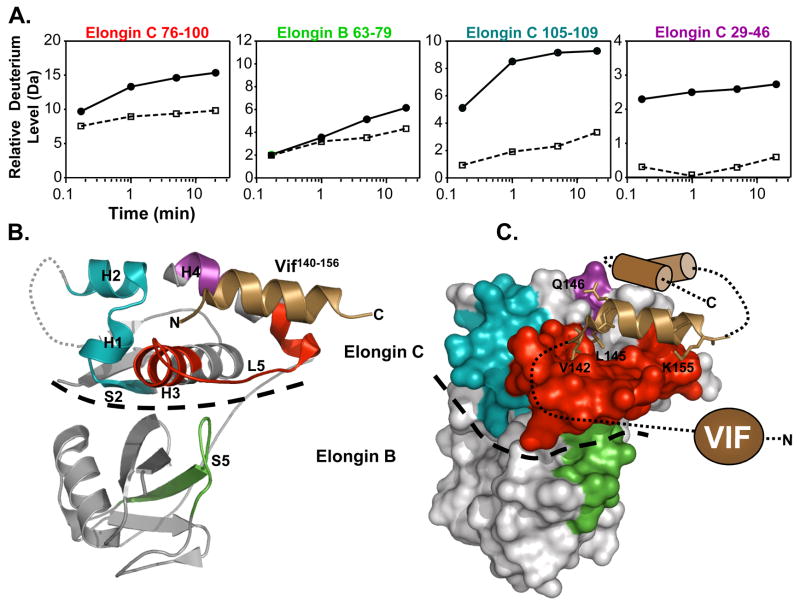
It has been shown previously with NMR that upon interaction with the VHL SOCS box containing peptide, Elongin C undergoes a conformational change.18 To determine if, how and where Vif might be able to stabilize the Elongin BC complex upon binding, Elongin BC was mixed with Vif and labeled with deuterium. The location of deuterium incorporation into Elongin BC was then determined following pepsin digestion and MS analysis. Deuterium uptake curves for Elongin BC peptides displaying changes upon incubation with Vif (Figure 5) show that several regions in Elongin C had marked differences in deuteration in the presence of Vif. These regions included residues 29–46 (S2, H1, L2, H2), 76–100 (H3, L5) and 105–109 (H4). Much less deuterium was incorporated into Elongin C in these regions when bound to full-length Vif. Elongin C H3, H4, and L5 make contacts with SOCS box containing proteins including Vif,12 see Figure 5B,C, and the changes in deuterium uptake lie within the binding pocket for Vif on the surface of Elongin C. Elongin B residues 63–79 (S5) also displayed a decrease in deuterium uptake upon interaction with Vif. The decreased dynamics of this region of Elongin B upon ligand binding could be explained by its proximity to the ligand binding region of Elongin C (H3, L5, and H4) which becomes stabilized by Vif binding (see Figure 5C). The decrease in deuterium uptake of S2, H1, and H2 of Elongin C is consistent with the idea that binding of SOCS box containing ligands alters the stability and conformation of Elongin C, and in the context of Vif binding, results in conformational changes that localize to the Vif:Elongin C interface.
Figure 5.
Location of conformational changes in Elongin B and C induced by HIV-1 Vif interaction. (A) Deuterium uptake curves for Elongin C and B peptic peptides that displayed changes in deuterium uptake upon interaction with Vif (all peptide data can be found in Figure S4). The solid lines represent the Elongin BC complex alone and the dotted lines represent the Elongin BC complex bound to Vif. The error for determination of the deuterium level was ±0.25Da. (B) The location of the changes mapped onto the ribbon diagram of the structure of the Vif140–156 :Elongin BC complex (PDB:3DCG) 12 and (C) onto a surface representation. The colors in panel A correspond to the colors in panels B,C (i.e. Elongin C 76–100, red; Elongin B 63–79, green; Elongin C 105–109, blue; Elongin C 29–46, purple). The grey sections indicate regions where there were no changes in deuterium uptake or the deuterium incorporation could not be monitored. The HIV-1 Vif peptide 140–156 is represented as the copper helix and the remaining structurally uncharacterized regions of Vif are depicted in cartoon form. A dotted line indicates the boundary between Elongin C and Elongin B.
Vif’s viral BC box undergoes a structural change in the presence of Elongin BC
It is unknown if Vif itself undergoes conformational change as a result of binding the Elongin BC complex. Reingewertz et al did not detect changes in the C-terminal domain of Vif upon incubation with an Elongin C peptide.16 Other folded SOCS box motifs have been observed in numerous structures with the Elongin BC complex 4–5 and it was reported that the SOCS box of SOCS 3 folded into the helical structure observed with NMR upon complexation with Elongin BC.4 If Vif were to gain structure or change conformation in the presence of the Elongin BC complex it would likely cause a difference in deuterium incorporation. In deuterium exchange experiments at the whole protein level, we were unable to detect major global conformational changes in Vif (peptic-peptide level changes in Vif will be reported elsewhere) upon interaction with the Elongin BC complex (see Figure S5). To further probe this interaction, in terms of potential structural changes in Vif, we utilized Vif135–158 in an HX MS experiment. HX MS data were obtained for Vif135–158 in the absence of (Figure 6A) or presence of (Figure 6B) Elongin BC. Note that after 10 seconds the mass increase of Vif135–158 alone did not change compared to the 1.5 hour time point. These data are indicative of an unstructured peptide and suggest that Vif135–158 does not contain any structural elements in its unbound state. When a calculation 35 is made for what one would expect to find in a totally unstructured peptide of the same sequence as Vif135–158 (gray line, Figure 6C), it closely resembles what is measured for Vif135–158 in solution (black line, Figure 6C). The red curve in Figure 6C is the deuterium uptake for Vif135–158 upon incubation with the Elongin BC complex based on the data in Figure 6B. Note initially there was a large decrease in deuterium uptake which became less apparent the longer the complex was labeled in 2H2O. This reduction in deuterium uptake is consistent with changes in solvent accessibility and hydrogen bonding in the Vif135–158, indicative of formation of structure in the presence of the Elongin BC complex (note that HX MS cannot be used to determine if the structure induced is a helix). While full length Vif does not appear to change its global conformation upon binding (Figure S5), the local region that is the main binding element (residues 135–158) does appear to change. A plausible explanation as to why changes are seen in Vif135–158 but not in full length Vif is that other changes in full length Vif conformation lead to increases in deuterium such that when viewed in total, the decreases in 135–158 are partially cancelled by increases elsewhere. The reduction in deuterium incorporation seen in Vif135–158 is too large to be explained merely by solvent protection, therefore implying that structural elements have formed in Vif135–158. Our results are consistent with known hypotheses indicating that structural elements (i.e., helices) must form in order for the SOCS box proteins to bind to Elongin BC.
Figure 6.
The Vif BC box becomes structured upon binding to Elongin BC. Deuteration of Vif135–158 in (A) the absence of the Elongin BC complex or (B) the presence of the Elongin BC complex. The + 5 charge state of Vif135–158 is shown. The deuterium labeling time is shown at the right and applies to spectra in both panels A and B (UN, undeuterated). The dotted lines are provided for visual guidance. (C) Deuterium uptake curves for data shown in panels A and B. The grey dotted line is the theoretical deuterium uptake for an unstructured peptide of the same sequence as calculated according to the Bai/Molday factors.35 The theoretical deuterium uptake was adjusted by subtracting the amide positions of the 6xHis tag due to the fast exchange properties of this sequence (i.e. all His residues should be undeuterated in Vif135–158 under the conditions used).46 The data for Vif135–158 have been corrected for back exchange (see Materials and Methods). Note the exchange properties of Vif135–158 in the absence of the Elongin BC complex (black line) nearly overlays the theoretical deuterium uptake for the Vif135–158 sequence if it did not contain any higher order structure (dotted line). Upon incubation with the Elongin BC complex (red line), Vif135–158 undergoes changes in deuterium uptake which are indicative of changes in solvent accessibility and hydrogen bonding.47 The Y-axis maximum is set to the number of backbone amide hydrogens in Vif135–158 capable of exchanging.
CONCLUSIONS
Previous studies provide evidence that HIV-1 Vif associates with the Elongin BC complex in vivo and that this interaction is essential for the degradation of APOBEC3 enzymes.8; 10 Our results show that full length Vif specifically binds to the Elongin BC complex in vitro and that this interaction does not require other cellular components. Figure 7 is a summary of the Vif:Elongin BC interaction as revealed by our results. In the unbound state, Elongin C is quite dynamic while Elongin B is quite stable. Upon binding to Vif, with a Kd of ~1.9 μM for full-length Vif, Elongin C is stabilized as is a small region of Elongin B that makes contact on the backside of the Elongin C residues that are stabilized. The Vif BC box folds while the remainder of the C-terminal domain of Vif likely stays disordered. To our knowledge Vif is the first SOCS box containing protein that has the ability to associate with the Elongin BC complex without the aid of other cellular factors or co-expression in vivo. Numerous cellular SOCS box containing proteins such as the Von Hippel Lindau (VHL) protein and SOCS 1-7 only bind to the Elongin BC complex when co-expressed in vivo.4; 36 The formation of the VHL:Elongin BC complex requires the chaperonin TRiC and SOCS 1-7 requires co-expression with Elongin BC to form a functional complex.15; 36 The ability of Vif to specifically interact with the Elongin BC complex independent of other cellular factors might be a unique property of Vif that allows it to more easily recruit components of the E3 ligase machinery.
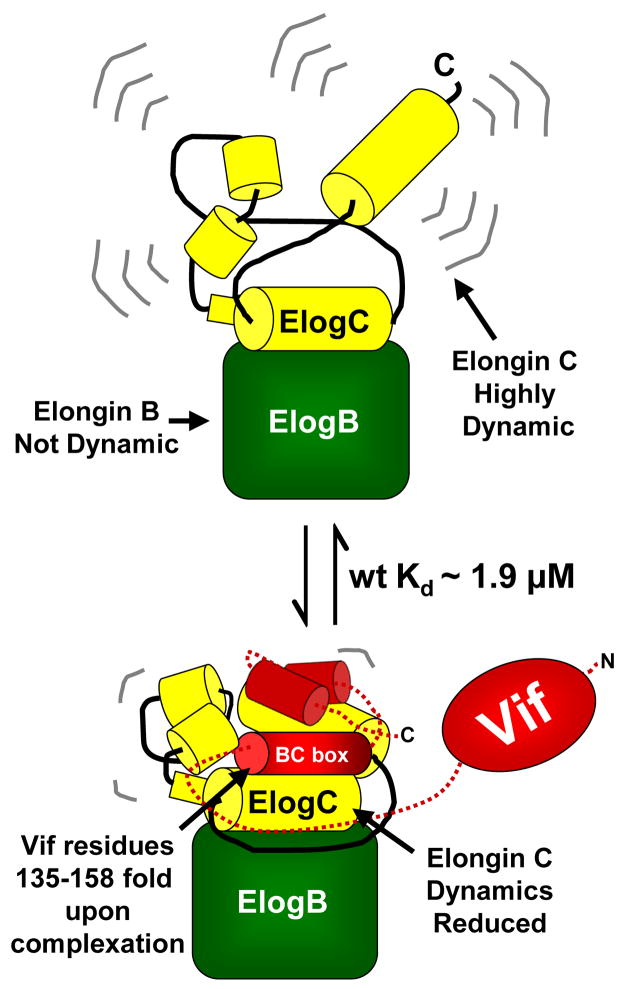
Figure 7.
Summary of the behavior of Elongin BC based on our results. In the Elongin BC complex alone, Elongin C is rapidly deuterated implying that the conformation is very flexible. Elongin B, in contrast, is much more difficult to deuterate, implying that it is more stable in solution. Unlike other SOCS boxes containing proteins,15 recombinant HIV-1 Vif interacts with the recombinant Elongin BC in vitro without additional factors. Full-length HIV-1 Vif (wt) binds to the Elongin BC complex with a Kd of ~1.9 μM and stabilizes Elongin C and a small region in Elongin B. The main region in Vif (the BC box residues 135–158) that is responsible for interacting with Elongin C folds upon binding.
We were unable to detect global conformational changes in full-length Vif upon incubation with Elongin BC. Deuterium incorporation into a Vif135–158 peptide changed dramatically upon Elongin BC incubation and suggests that this region folds (into a helical structure according to similarity with other SOCS systems) upon binding and likely does so in the full length protein. Such an event would be consistent with the structures of SOCS box containing proteins seen in numerous structures of the Elongin BC complex, including that observed for SOCS3.3 The physiological relevance of the low micro-molar dissociation constants reported in this in vitro study and that of Bergeron et al23 can likely be attributed to several factors. Additional stabilization of the Vif:Elongin BC complex could possibly arise from cellular factors present in vivo that were absent in vitro such as Cullin 5 and/or zinc. The dynamic and unstructured nature of the C-terminal tail of Vif could also explain the moderate affinity of full length Vif, compared to shorter Vif constructs, for Elongin BC. The entropic energy costs of folding would be less significant in shorter Vif constructs and the possible steric effects of the additional residues present in full-length Vif would be greater than in Vif135–158. The necessity of a high affinity interaction between Vif and the Elongin BC complex might be partially negated due to the additional mechanisms in which Vif circumvents the antiviral activities of the APOBEC3 proteins. Vif has been shown to regulate APOBEC3G protein levels by directly inhibiting APOBEC3G mRNA translation 37 and promoting the formation of APOBEC3G high molecular weight complexes.38 Regardless of the differences in dissociation constants reported in the literature for the Vif:Elongin BC interaction, the ability of Vif to specifically interact with the Elongin BC complex independent of other cellular factors is an unique and promising target for anti-HIV-1 drug design. The ability of our HX MS assay to detect Vif:Elongin BC binding provides an unique tool to screen for Vif:Elongin BC inhibitors.
MATERIALS and METHODS
DNA Constructs and protein purification
Full length HIV-1 Vif (HXB2), VifL145A, VifΔSLQ (SLQ to AAA), and Vif1–141 were over-expressed and purified using the pET28b vector as described.39 The HIV-1 Vif pET28b codon optimized construct was a gift from Dana Gabuzda at Harvard Medical School. VifL145A, ΔSLQ, and 1–141 were constructed using Lightning Quick Change® (Stratagene, La Jolla, CA) site directed mutagenesis from the HIV-1 Vif (HXB2) template. Human Elongin B and C (residues 17–122) were co-expressed using the pACYCDUET-ElCB vector in BL21(DE3).3 The pACYCDUET-ElCB vector was a gift from Alex Bullock and Stefan Knapp at the Structural Genomics Consortium, University of Oxford. The Elongin BC complex was purified by anion-exchange on a HiTrap QHP column followed by size exclusion chromatography on a Superose 12 10/300 column (GE Healthcare). All purified proteins were dialyzed into Vif buffer (20 mM MOPS, 150 mM NaCl, 1mM DTT, 10% glycerol, pH 7.0) and stored at −80 ºC. Vif135–158 (HHHHHH135YQAGHNKVGSLQYLALAALITPKK158) was designed from the HXB2 Vif viral BC box sequence with a 6xHis affinity tag and synthesized by Gen Script (Piscataway, NJ). The peptide (>98% purity) was dissolved directly into Vif buffer and stored at −80 °C. All concentration values in all experiments were determined with the Bradford assay (Bio-Rad protein Assay, Hercules, CA).40 All proteins masses were verified by mass spectrometry and are shown in the Supplementary Material, Figure S1.
Pull-down analysis of the HIV-1 Vif:Elongin BC interaction
For pull down experiments between Vif constructs and the Elongin BC complex, 1 nmol of each Vif construct was immobilized on Ni-NTA agarose. The Vif immobilized Ni-NTA agarose was then incubated with 3 nmol of the Elongin BC complex for 30 minutes at 4 °C. After incubation each reaction was washed 7 times with 1 mL each of Vif buffer. 20 μLs of each reaction was then analyzed with SDS-PAGE and visualized with coomassie staining. Relevant bands were then quantified with GE Healthcare (Piscataway, NJ) Image Quant TL 1D software. All experiments were conducted at least twice.
Intact protein HX MS
The Elongin BC complex (4.00 μM) was incubated with or without a four-fold molar excess of full-length wt Vif (16 μM) at 4 °C for 30 minutes. Protein mixtures were then diluted 10-fold with 20 mM MOPS, 150 mM NaCl, 1 mM DTT (pD 7.0), 2H2O at 20 °C and allowed to incubate for various times. The concentrations of Elongin BC and Vif after dilution into 2H2O were 0.36 μM and 1.45 μM, respectively. The percentage of Elongin BC bound to Vif was 41% during labeling reactions based on a Kd of 1.9 μM. The labeling reaction was quenched by adjusting an aliquot to pH 2.6 with 0.8 M guanidine hydrochloride, 0.8 % formic acid. Quenched (1.1 mL total volume, 200 pmols Elongin BC) samples were then immediately injected into a Shimadzu SCL-10A VP HPLC flowing at 50 μL/min coupled to a Waters LCT premier mass spectrometer with a standard electrospray interface. Proteins were eluted directly into the mass spectrometer with a gradient of 8–98 % acetonitrile in eight minutes. Both mobile phases contained 0.05% formic acid. The injector, column (Alltech in-line refillable analytical guard column (Grace, Deerfield, IL) packed with POROS 20-R2 media, PerSeptive Biosystems), and all associated tubing were kept at 0 °C to minimize back exchange.25 Deuterium levels were not corrected for back exchange and are therefore reported as relative deuterium levels.20 Intact mass spectra were deconvoluted using the software MagTran.41 The intact protein relative deuterium uptake was calculated by subtracting the centroid mass of undeuterated protein from the centroid mass of deuterium labeled protein.
HX MS titrations
For Elongin BC titrations with HIV-1 Vif, each Vif construct was titrated into a fixed concentration of Elongin BC (3.22 μM). The Vif:Elongin BC mixtures were incubated at 4 ºC for 30 minutes before labeling. The mixtures were then diluted with a 10-fold excess of 20 mM MOPS, 150 mM NaCl, 1 mM DTT (pD 7.0), 2H2O at 20 ºC and labeling allowed to proceed for 10 seconds before quenching. After the quench, protein mixtures were analyzed as stated above. The percentage of Elongin C bound and unbound was determined by fitting the area under a charge state of Elongin C with a Gaussian distribution using the PeakFit® program (Systat, San Jose, CA). The percentage of Elongin C bound was then plotted versus Vif concentration (μM) and the data were fit using a sigmoidal equation with Sigma Plot® software (Systat Software Inc, San Jose, CA). The reported Kd (Vif concentration that resulted in 50% Elongin C bound) was an average from duplicate experiments. No binding was detected between Vif1–141 and the Elongin BC complex.
Peptide-level HX MS
Hydrogen exchange was preformed with a 10-fold dilution into 2H2O and then quenched as stated above . Quenched samples (110 μl total volume, 20 pmols of Elongin BC) was digested, desalted, and separated online using a custom Waters nanoACUITY UPLC system.42 Online digestion was performed in 0.05 % formic acid at 20 °C at a flow rate of 50 μL/min through a 2.1 mm x 50 mm stainless steel column packed with pepsin immobilized on POROS-20AL beads (PerSeptive Biosystems).43 Peptic peptides were trapped on an AQUITY UPLC BEH C8 1.7 μm peptide trap at 0 °C and desalted for 3 minutes before separation on an AQUITY UPLC BEH C8 1.7 μm 1.0 X 100 mm column (Waters, Milford, MA) at 0 °C with a flow rate of 40 μL/min. A 6 minute 6–40 % acetonitrile gradient with 0.05 % formic acid (pH 2.6) was used to elute the peptides directly into a Waters QToF premier mass spectrometer with standard electrospray interface. Continuous lock mass correction was carried out using Glu-fibrinogen peptide and peptic peptides were identified using MSE and Waters IdentityE software.44 The deuterium uptake for each peptide was determined using the excel based software program HX-Express 45 by subtracting the centroid mass of each undeuterated peptide from the centroid mass of deuterium labeled peptides. No back exchange correction was applied (except for that noted in the next paragraph) and all values are therefore reported as relative.20 All experiments were conducted at least twice.
For experiments with the Elongin BC complex and Vif135–158, Vif135–158 (4.00 μM) was incubated with or without a four-fold molar excess of the Elongin BC complex (16 μM). The percentage of Vif135–158 bound to the Elongin BC complex during equilibration was calculated to be 83% during the labeling reaction based on a Kd of 0.23 μM. Deuterium uptake values for Vif135–158 were adjusted for back exchange in order to make a comparison with the predicted theoretical exchange. For the fully deuterated control, Vif135–158 was incubated at 37 °C for 7 hours in a 10-fold excess of 2H2O before quenching and analysis as above. For back exchange correction equation 1 25 was used.
| (Eq.1) |
D0 is the back exchange adjusted amount of deuterium atoms incorporated into a protein after incubation in 2H2O. <m>, <m0%>, and <m100%> are the observed masses of the protein partially deuterated, undeuterated, and fully deuterated, respectively. N is the number of exchangeable amide positions in a protein. N was adjusted for by subtracting the amide positions of the 6xHis tag due to the fast exchange properties of this sequence.46
Supplementary Material
Acknowledgments
We are pleased to thank Dana Gabuzda from Harvard Medical School for the HIV-1 Vif pET28b codon optimized construct, and Alex Bullock and Stefan Knapp at the Structural Genomics Consortium, University of Oxford for the pACYCDUET-ElCB vector. This work was supported in part by the National Institutes of Health (R01-GM070590 and R01-GM086507) and a research collaboration with the Waters Corporation. This is contribution 969 from the Barnett Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Duan DR, Pause A, Burgess WH, Aso T, Chen DY, Garrett KP, Conaway RC, Conaway JW, Linehan WM, Klausner RD. Inhibition of transcription elongation by the VHL tumor suppressor protein. Science. 1995;269:1402–6. doi: 10.1126/science.7660122. [DOI] [PubMed] [Google Scholar]
- 2.Conaway JW, Kamura T, Conaway RC. The Elongin BC complex and the von Hippel-Lindau tumor suppressor protein. Biochim Biophys Acta. 1998;1377:M49–54. doi: 10.1016/s0304-419x(97)00035-8. [DOI] [PubMed] [Google Scholar]
- 3.Bullock AN, Debreczeni JE, Edwards AM, Sundstrom M, Knapp S. Crystal structure of the SOCS2-elongin C-elongin B complex defines a prototypical SOCS box ubiquitin ligase. Proc Natl Acad Sci U S A. 2006;103:7637–42. doi: 10.1073/pnas.0601638103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Babon JJ, Sabo JK, Soetopo A, Yao S, Bailey MF, Zhang JG, Nicola NA, Norton RS. The SOCS box domain of SOCS3: structure and interaction with the elonginBC-cullin5 ubiquitin ligase. J Mol Biol. 2008;381:928–40. doi: 10.1016/j.jmb.2008.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bullock AN, Rodriguez MC, Debreczeni JE, Songyang Z, Knapp S. Structure of the SOCS4-ElonginB/C complex reveals a distinct SOCS box interface and the molecular basis for SOCS-dependent EGFR degradation. Structure. 2007;15:1493–504. doi: 10.1016/j.str.2007.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mehle A, Goncalves J, Santa-Marta M, McPike M, Gabuzda D. Phosphorylation of a novel SOCS-box regulates assembly of the HIV-1 Vif-Cul5 complex that promotes APOBEC3G degradation. Genes Dev. 2004;18:2861–6. doi: 10.1101/gad.1249904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fang L, Landau NR. Analysis of Vif-induced APOBEC3G degradation using an alpha-complementation assay. Virology. 2007;359:162–9. doi: 10.1016/j.virol.2006.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu Y, Xiao Z, Ehrlich ES, Yu X, Yu XF. Selective assembly of HIV-1 Vif-Cul5-ElonginB-ElonginC E3 ubiquitin ligase complex through a novel SOCS box and upstream cysteines. Genes Dev. 2004;18:2867–72. doi: 10.1101/gad.1250204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–50. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- 10.Kobayashi M, Takaori-Kondo A, Miyauchi Y, Iwai K, Uchiyama T. Ubiquitination of APOBEC3G by an HIV-1 Vif-Cullin5-Elongin B-Elongin C complex is essential for Vif function. J Biol Chem. 2005;280:18573–8. doi: 10.1074/jbc.C500082200. [DOI] [PubMed] [Google Scholar]
- 11.Mehle A, Strack B, Ancuta P, Zhang C, McPike M, Gabuzda D. Vif overcomes the innate antiviral activity of APOBEC3G by promoting its degradation in the ubiquitin-proteasome pathway. J Biol Chem. 2004;279:7792–8. doi: 10.1074/jbc.M313093200. [DOI] [PubMed] [Google Scholar]
- 12.Stanley BJ, Ehrlich ES, Short L, Yu Y, Xiao Z, Yu XF, Xiong Y. Structural insight into the human immunodeficiency virus Vif SOCS box and its role in human E3 ubiquitin ligase assembly. J Virol. 2008;82:8656–63. doi: 10.1128/JVI.00767-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shao Q, Wang Y, Hildreth JE, Liu B. Polyubiquitination of APOBEC3G is essential for its degradation by HIV-1 Vif. J Virol. 84:4840–4. doi: 10.1128/JVI.01911-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simon JH, Sheehy AM, Carpenter EA, Fouchier RA, Malim MH. Mutational analysis of the human immunodeficiency virus type 1 Vif protein. J Virol. 1999;73:2675–81. doi: 10.1128/jvi.73.4.2675-2681.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Babon JJ, Sabo JK, Zhang JG, Nicola NA, Norton RS. The SOCS box encodes a hierarchy of affinities for Cullin5: implications for ubiquitin ligase formation and cytokine signalling suppression. J Mol Biol. 2009;387:162–74. doi: 10.1016/j.jmb.2009.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reingewertz TH, Benyamini H, Lebendiker M, Shalev DE, Friedler A. The C-terminal domain of the HIV-1 Vif protein is natively unfolded in its unbound state. Protein Eng Des Sel. 2009;22:281–7. doi: 10.1093/protein/gzp004. [DOI] [PubMed] [Google Scholar]
- 17.Reingewertz TH, Shalev DE, Friedler A. Structural Disorder in the HIV-1 Vif Protein and Interaction-Dependent Gain of Structure. Protein Pept Lett. 2010 doi: 10.2174/092986610791498876. in press. [DOI] [PubMed] [Google Scholar]
- 18.Botuyan MV, Koth CM, Mer G, Chakrabartty A, Conaway JW, Conaway RC, Edwards AM, Arrowsmith CH, Chazin WJ. Binding of elongin A or a von Hippel-Lindau peptide stabilizes the structure of yeast elongin C. Proc Natl Acad Sci U S A. 1999;96:9033–8. doi: 10.1073/pnas.96.16.9033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marcsisin SR, Engen JR. Hydrogen exchange mass spectrometry: what is it and what can it tell us? Anal Bioanal Chem. 2010;397:967–72. doi: 10.1007/s00216-010-3556-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wales TE, Engen JR. Hydrogen exchange mass spectrometry for the analysis of protein dynamics. Mass Spectrom Rev. 2006;25:158–70. doi: 10.1002/mas.20064. [DOI] [PubMed] [Google Scholar]
- 21.Engen JR. Analysis of protein conformation and dynamics by hydrogen/deuterium exchange MS. Anal Chem. 2009;81:7870–5. doi: 10.1021/ac901154s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wolfe LS, Stanley BJ, Liu C, Eliason WK, Xiong Y. Dissection of HIV Vif interaction with the human E3 ubiquitin ligase. J Virol. 2010 doi: 10.1128/JVI.00031–10. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bergeron JR, Huthoff H, Veselkov DA, Beavil RL, Simpson PJ, Matthews SJ, Malim MH, Sanderson MR. The SOCS-box of HIV-1 Vif interacts with ElonginBC by induced-folding to recruit its Cul5-containing ubiquitin ligase complex. PLoS Pathog. 6:e1000925. doi: 10.1371/journal.ppat.1000925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Engen JR. Analysis of protein complexes with hydrogen exchange and mass spectrometry. Analyst. 2003;128:623–8. doi: 10.1039/b212800b. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Z, Smith DL. Determination of amide hydrogen exchange by mass spectrometry: a new tool for protein structure elucidation. Protein Sci. 1993;2:522–31. doi: 10.1002/pro.5560020404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mayne L, Paterson Y, Cerasoli D, Englander SW. Effect of antibody binding on protein motions studied by hydrogen-exchange labeling and two-dimensional NMR. Biochemistry. 1992;31:10678–10685. doi: 10.1021/bi00159a006. [DOI] [PubMed] [Google Scholar]
- 27.Zhang J, Adrian FJ, Jahnke W, Cowan-Jacob SW, Li AG, Iacob RE, Sim T, Powers J, Dierks C, Sun F, Guo GR, Ding Q, Okram B, Choi Y, Wojciechowski A, Deng X, Liu G, Fendrich G, Strauss A, Vajpai N, Grzesiek S, Tuntland T, Liu Y, Bursulaya B, Azam M, Manley PW, Engen JR, Daley GQ, Warmuth M, Gray NS. Targeting Bcr-Abl by combining allosteric with ATP-binding-site inhibitors. Nature. 2010;463:501–6. doi: 10.1038/nature08675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Botuyan MV, Mer G, Yi GS, Koth CM, Case DA, Edwards AM, Chazin WJ, Arrowsmith CH. Solution structure and dynamics of yeast elongin C in complex with a von Hippel-Lindau peptide. J Mol Biol. 2001;312:177–86. doi: 10.1006/jmbi.2001.4938. [DOI] [PubMed] [Google Scholar]
- 29.Buchberger A, Howard MJ, Freund SM, Proctor M, Butler PJ, Fersht AR, Bycroft M. Biophysical characterization of elongin C from Saccharomyces cerevisiae. Biochemistry. 2000;39:11137–46. doi: 10.1021/bi0008231. [DOI] [PubMed] [Google Scholar]
- 30.Brower CS, Shilatifard A, Mather T, Kamura T, Takagi Y, Haque D, Treharne A, Foundling SI, Conaway JW, Conaway RC. The elongin B ubiquitin homology domain. Identification of Elongin B sequences important for interaction with Elongin C. J Biol Chem. 1999;274:13629–36. doi: 10.1074/jbc.274.19.13629. [DOI] [PubMed] [Google Scholar]
- 31.Hon WC, Wilson MI, Harlos K, Claridge TD, Schofield CJ, Pugh CW, Maxwell PH, Ratcliffe PJ, Stuart DI, Jones EY. Structural basis for the recognition of hydroxyproline in HIF-1 alpha by pVHL. Nature. 2002;417:975–8. doi: 10.1038/nature00767. [DOI] [PubMed] [Google Scholar]
- 32.Auclair JR, Green KM, Shandilya S, Evans JE, Somasundaran M, Schiffer CA. Mass spectrometry analysis of HIV-1 Vif reveals an increase in ordered structure upon oligomerization in regions necessary for viral infectivity. Proteins. 2007;69:270–84. doi: 10.1002/prot.21471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang B, Gao L, Li L, Lu Z, Fan X, Patel CA, Pomerantz RJ, DuBois GC, Zhang H. Potent suppression of viral infectivity by the peptides that inhibit multimerization of human immunodeficiency virus type 1 (HIV-1) Vif proteins. J Biol Chem. 2003;278:6596–602. doi: 10.1074/jbc.M210164200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang S, Sun Y, Zhang H. The multimerization of human immunodeficiency virus type I Vif protein: a requirement for Vif function in the viral life cycle. J Biol Chem. 2001;276:4889–93. doi: 10.1074/jbc.M004895200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bai Y, Milne JS, Mayne L, Englander SW. Primary structure effects on peptide group hydrogen exchange. Proteins. 1993;17:75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feldman DE, Thulasiraman V, Ferreyra RG, Frydman J. Formation of the VHL-elongin BC tumor suppressor complex is mediated by the chaperonin TRiC. Mol Cell. 1999;4:1051–61. doi: 10.1016/s1097-2765(00)80233-6. [DOI] [PubMed] [Google Scholar]
- 37.Mercenne G, Bernacchi S, Richer D, Bec G, Henriet S, Paillart JC, Marquet R. HIV-1 Vif binds to APOBEC3G mRNA and inhibits its translation. Nucleic Acids Res. 2010;38:633–46. doi: 10.1093/nar/gkp1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goila-Gaur R, Khan MA, Miyagi E, Kao S, Opi S, Takeuchi H, Strebel K. HIV-1 Vif promotes the formation of high molecular mass APOBEC3G complexes. Virology. 2008;372:136–46. doi: 10.1016/j.virol.2007.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang X, Goncalves J, Gabuzda D. Phosphorylation of Vif and its role in HIV-1 replication. J Biol Chem. 1996;271:10121–9. doi: 10.1074/jbc.271.17.10121. [DOI] [PubMed] [Google Scholar]
- 40.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–54. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 41.Zhang Z, Marshall AG. A universal algorithm for fast and automated charge state deconvolution of electrospray mass-to-charge ratio spectra. J Am Soc Mass Spectrom. 1998;9:225–33. doi: 10.1016/S1044-0305(97)00284-5. [DOI] [PubMed] [Google Scholar]
- 42.Wales TE, Fadgen KE, Gerhardt GC, Engen JR. High-speed and high-resolution UPLC separation at zero degrees Celsius. Anal Chem. 2008;80:6815–20. doi: 10.1021/ac8008862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang L, Pan H, Smith DL. Hydrogen exchange-mass spectrometry: optimization of digestion conditions. Mol Cell Proteomics. 2002;1:132–8. doi: 10.1074/mcp.m100009-mcp200. [DOI] [PubMed] [Google Scholar]
- 44.Silva JC, Denny R, Dorschel C, Gorenstein MV, Li GZ, Richardson K, Wall D, Geromanos SJ. Simultaneous qualitative and quantitative analysis of the Escherichia coli proteome: a sweet tale. Mol Cell Proteomics. 2006;5:589–607. doi: 10.1074/mcp.M500321-MCP200. [DOI] [PubMed] [Google Scholar]
- 45.Weis DD, Engen JR, Kass IJ. Semi-automated data processing of hydrogen exchange mass spectra using HX-Express. J Am Soc Mass Spectrom. 2006;17:1700–3. doi: 10.1016/j.jasms.2006.07.025. [DOI] [PubMed] [Google Scholar]
- 46.Bache N, Rand KD, Roepstorff P, Ploug M, Jorgensen TJ. Hydrogen atom scrambling in selectively labeled anionic peptides upon collisional activation by MALDI tandem time-of-flight mass spectrometry. J Am Soc Mass Spectrom. 2008;19:1719–25. doi: 10.1016/j.jasms.2008.05.021. [DOI] [PubMed] [Google Scholar]
- 47.Morgan CR, Engen JR. Investigating solution-phase protein structure and dynamics by hydrogen exchange mass spectrometry. Curr Protoc Protein Sci. 2009;Chapter 17(Unit 17):6, 1–17. doi: 10.1002/0471140864.ps1706s58. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.