Abstract
Seventy-three chronic hepatitis B patients who had either hepatitis B e antigen (HBeAg) seroconversion (group I) or HBeAg-negative disease (group II) were studied. HBV DNA levels at HBeAg seroconversion (group I) and at initial visits (group II) were significantly lower among patients who were persistently negative for HBeAg than among those who underwent HBeAg reversion.
In chronic hepatitis B, hepatitis B e antigen (HBeAg)-negative inactive disease (normal liver enzymes) has been shown to be associated with low risk of liver-related complications (7, 8, 11, 12), whereas HBeAg reversion is associated with increased risk of liver-related complications, hepatocellular carcinoma, and mortality (8, 9, 11). This study aimed to identify a level of viral load that can predict sustained disease remission after HBeAg seroconversion and in HBeAg-negative patients in a longitudinal follow-up.
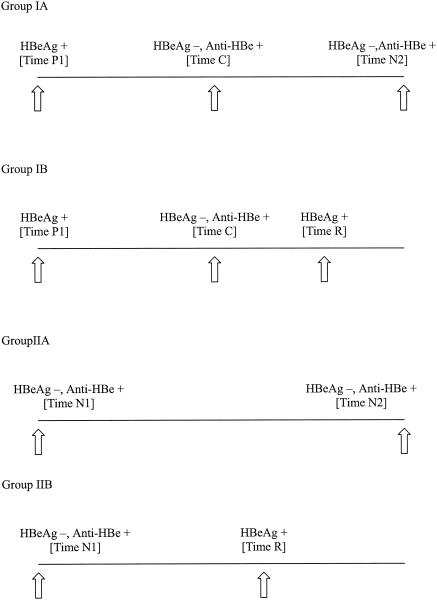
Residual serum samples from treatment-naïve chronic hepatitis B patients were studied (2). The patients were followed up at 6-month intervals or more frequently as clinically indicated. Group I patients were positive for HBeAg on their initial visits and developed HBeAg seroconversion. Group IA patients had sustained HBeAg seroconversion for at least 6 months with normal alanine transaminase (ALT) levels, while group IB patients had reversion of HBeAg. Group II patients were negative for HBeAg and positive for anti-HBe at their initial visits. Group IIA patients were negative for HBeAg, with normal ALT levels throughout the follow-up, while Group IIB patients had HBeAg reversion.
For group I patients, HBV DNA was measured at their initial visits (time P1), at the time HBeAg first became negative (HBeAg seroconversion; time C), at the last visit (group IA; time N2), and at the time of HBeAg reversion (group IB; time R) (Fig. 1). For group II patients, HBV DNA was measured at their initial visits (time N1), at the last visit (group IIA; time N2), and at HBeAg reversion (group IIB; time R).
FIG. 1.
Time points of serum sampling for HBV DNA testing. Group IA, sustained HBeAg seroconversion; group IB, HBeAg reversion after seroconversion; group IIA, persistently HBeAg-negative inactive disease; group IIB, HBeAg reversion in HBeAg-negative patients. +, positive; −, negative.
Serum HBV DNA was quantified by the TaqMan real-time PCR system (PE Biosystems, Foster City, Calif.) as described previously (4, 5, 16). A standard curve was generated by serial 10-fold dilution of EUROHEP genotype D HBV standard (from K. H. Heerman, University of Goettingen, Goettingen, Germany), which contained 2.7 × 109 viral copies per ml. The range of HBV DNA detection was from 102 to 109 copies/ml. HBV genotyping was performed in the initial serum sample of each patient by restriction fragment length polymorphism as described previously (3, 5, 9).
HBV DNA was logarithmically transformed to normal distribution and expressed as the mean ± standard deviation. Variables were analyzed by a logistic regression model. Statistical significance was taken as a P value of <0.05.
The patients in group IA were younger than those in group IB, and the patients in group IIA were younger and had significantly lower initial ALT level than those in group IIB (Table 1). Other characteristics were comparable between the paired patient groups.
TABLE 1.
Demographic and clinical characteristics of patients
| Characteristic | Valuea
|
|||
|---|---|---|---|---|
| Group IA | Group IB | Group IIA | Group IIB | |
| n | 20 | 13 | 29 | 11 |
| Male/female ratio | 11:9 | 11:2 | 12:17 | 8:3 |
| Age (yr)b | 29 (12-39) | 39 (13-68) | 41 (21-65) | 49 (37-67) |
| Initial ALTb | 67 (18-455) | 68 (25-294) | 30 (16-58) | 52 (32-161) |
| Follow-up duration (wk)b | 159 (102-208) | 180 (49-704) | 186 (64-226) | 165 (49-705) |
| Genotype B/C ratioc | 6:13 | 3:10 | 4:22 | 2:8 |
Group IA, sustained HBeAg seroconversion; group IB, HBeAg reversion after seroconversion; group IIA, persistently HBeAg-negative inactive disease; group IIB, HBeAg reversion in HBeAg-negative patients. P = 0.03 (group IA versus group IB) and P = 0.04 (group IIA versus group IIB) for age; P = 0.003 (group IIA versus group IIB) for initial ALT; for all other comparisons between groups IA and IB and groups IIA and IIB, P > 0.05.
Median (range).
1, 3, and 1 patients in groups IA, IIA, and IIB, respectively, had negative PCR on genotyping.
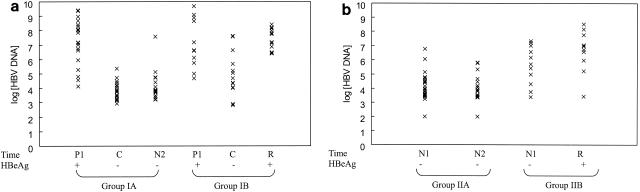
The patients in groups IA and IB were positive for HBeAg for 70 (range, 3 to 131) weeks and 47 (range, 4 to 153) weeks before HBeAg seroconversion (P = 0.82). Patients who had sustained HBeAg seroconversion (group IA) were followed up for 89 (range, 28 to 155) weeks postseroconversion. HBeAg reversion occurred at 32 (range, 5 to 140) weeks after seroconversion in group IB. The log10 HBV DNA levels at initial visits (time P1) were comparable between the patients in groups IA (7.23 ± 1.50) and IB (7.24 ± 1.63) (P = 1.0) (Fig. 2a). The log10 HBV DNA levels at HBeAg seroconversion (time C) were significantly lower among the patients in group IA (3.87 ± 0.61) than among those in group IB (4.99 ± 1.55) (P = 0.007). The log10 HBV DNA levels at HBeAg seroconversion (time C) among patients in group IA were comparable to those at their last visit (time N2) (4.11 ± 0.98) (P = 0.3). At HBeAg reversion (time R) among patients in group IB, the log10 HBV DNA level (7.39 ± 0.75) rose to the level at the initial visit (time P1) (P = 0.8). Using the log10 HBV DNA level at time C to predict HBeAg reversion, the area under the receiver operator characteristic curve was 0.73 (95% confidence interval, 0.53, 0.93; P = 0.03), and the likelihood ratios of the log10 HBV DNA level being <4 and >5 were 0.3 and 9.2, respectively (Table 2).
FIG. 2.
Point plots of log10 HBV DNA levels at various time points among patients in group I (a) and group II (b). Each plot represents the log10 HBV DNA level of a patient at a particular time point. (a) Times: P1, initial visit when HBeAg was positive; C, time of HBeAg seroconversion; N2, last visit for patients who had sustained HBeAg seroconversion; R, HBeAg reversion. (b) Times: N1, initial visit; N2, last visit for patients who had sustained negative HBeAg; R, HBeAg reversion. +, positive; −, negative.
TABLE 2.
Likelihood ratios of different log10 HBV DNA levels to predict HBeAg reversion
| Group (time)a | Log10 HBV DNA | No. (%) of patients in subgroup:
|
Likelihood ratio | |
|---|---|---|---|---|
| A | B | |||
| I (C) | <4 | 11 (55.0) | 2 (15.4) | 0.3 |
| 4-5 | 8 (40.0) | 5 (38.5) | 1.0 | |
| >5 | 1 (5.0) | 6 (46.2) | 9.2 | |
| II (N1) | <4 | 19 (65.5) | 2 (18.2) | 0.3 |
| 4-5 | 7 (24.1) | 2 (18.2) | 0.8 | |
| >5 | 3 (10.3) | 7 (53.8) | 5.2 | |
Group IA, sustained HBeAg seroconversion; group IB, HBeAg reversion after seroconversion; group IIA, persistently HBeAg-negative inactive disease; group IIB, HBeAg reversion in HBeAg-negative patients; time C, first disappearance of HBeAg in HBeAg-positive patients; time N1, initial visit of HBeAg-negative patients.
The log10 HBV DNA levels at initial visits (time N1) were lower among patients in group IIA (3.84 ± 1.10) than among those in group IIB (5.62 ± 1.39) (P = 0.0001) (Fig. 2b). The log10 HBV DNA levels at the initial (time N1) and last (time N2) (3.33 ± 1.18) visits were comparable among the patients in group IIA (P = 0.1). HBeAg reversion occurred at 48 (range, 16 to 129) weeks in group IIB. There was an increase in log10 HBV DNA levels at HBeAg reversion (time R) among patients in group IIB (6.65 ± 1.43), but the difference did not reach statistical significance compared to that at the initial visit (time N1) (P = 0.3). Using the log10 HBV DNA at time N1 to predict HBeAg reversion, the area under the receiver operator characteristic curve was 0.83 (95% confidence interval, 0.67, 0.98; P = 0.002), and the likelihood ratios of the log10 HBV DNA level being <4 and >5 were 0.3 and 5.2, respectively (Table 2). All patients in group IIB who had elevated initial ALT levels had log10 HBV DNA levels of >5.
The results of this study showed that a significant drop in HBV DNA was required for HBeAg seroconversion. An HBV DNA level below 104 copies/ml was predictive of inactive liver disease among patients undergoing HBeAg seroconversion and in HBeAg-negative patients, as reported in Europe (10, 14, 15). On the other hand, an HBV DNA level above 105 copies/ml was predictive of HBeAg reversion and viral recurrence, in line with the findings of anther Asian study (6). This suggested that insufficient viral suppression in HBeAg-negative patients was associated with a higher risk of viral recurrence. Some HBeAg-negative patients have high HBV DNA levels and HBeAg reversion despite persistently normal ALT levels. This echoed the previous findings that a normal ALT level alone was not an accurate indicator of inactive disease (1, 3, 17, 18).
Substantial overlap in HBV DNA levels did exist between patients who were in remission versus those who had HBeAg reversion, and an HBV DNA level between 104 to 105 copies/ml was a grey zone to predict disease activity. Although HBV DNA levels fluctuated with time, the magnitude of fluctuation in HBeAg-negative patients was limited, as illustrated by the similar HBV DNA levels at times C and N2 in group IA and at times N1 and N2 in group IIA.
In summary, measurement of HBV DNA after HBeAg seroconversion and in HBeAg-negative patients is useful to assess the risk of HBeAg reversion. Our results conform to the National Institutes of Health recommendation that an HBV DNA level higher than 105 copies/ml is associated with active liver disease (13). More sensitive HBV DNA assays are needed to detect lower HBV DNA levels for identification of inactive carriers.
Acknowledgments
This study was supported by an RGC Earmarked Research Grant—Direct Allocation, Hong Kong (reference number 2002.1.023).
REFERENCES
- 1.Chan, H. L. Y., Y. Hui, N. W. Y. Leung, J. Y. L. Ching, F. K. L. Chan, and J. J. Y. Sung. 2000. Risk factors for active liver disease in HBeAg-negative chronic hepatitis B virus (HBV) infected patients. Am. J. Gastroenterol. 95:3547-3551. [DOI] [PubMed] [Google Scholar]
- 2.Chan, H. L. Y., N. W. Y. Leung, M. Hussain, M. L. Wong, and A. S. F. Lok. 2000. Hepatitis B e antigen-negative chronic hepatitis B in Hong Kong. Hepatology 31:763-768. [DOI] [PubMed] [Google Scholar]
- 3.Chan, H. L. Y., S. W. C. Tsang, C. T. Liew, C. H. Tse, M. L. Wong, J. Y. L. Ching, N. W. Y. Leung, J. S. L. Tam, and J. J. Y. Sung. 2002. Viral genotype and hepatitis B virus DNA levels are correlated with histologic liver damage in HBeAg-negative chronic hepatitis B virus infection. Am. J. Gastroenterol. 97:406-412. [DOI] [PubMed] [Google Scholar]
- 4.Chan H. L. Y., A. K. K. Chui, W. Y. Lau, F. K. L. Chan, M. L. Wong, C. H. Tse, A. R. N. Rao, J. Wong, and J. J. Y. Sung. 2002. Factors associated with viral breakthrough in lamivudine monoprophylaxis of hepatitis B virus recurrence after liver transplantation. J. Med. Virol. 68:182-187. [DOI] [PubMed] [Google Scholar]
- 5.Chan, H. L. Y., M. L. Wong, A. Y. Hui, L. C. T. Hung, F. K. L. Chan, and J. J. Y. Sung. 2003. Hepatitis B virus genotype C takes a more aggressive disease course than hepatitis B virus genotype B in hepatitis B e antigen-positive patients. J. Clin. Microbiol. 41:1277-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chu, C. J., M. Hussain, and A. S. F. Lok. 2002. Quantitative serum HBV DNA levels during different stages of chronic hepatitis B infection. Hepatology 36:1408-1415. [DOI] [PubMed] [Google Scholar]
- 7.Di Marco, V., O. L. Iacono, C. Camma, A. Vaccaro, M. Giunta, G. Martorana, P. Fuschi, P. Almasio, and A. Craxi. 1999. The long-term course of chronic hepatitis B. Hepatology 30:157-164. [DOI] [PubMed] [Google Scholar]
- 8.Hsu, Y. S., R. N. Chien, C. T. Yeh, I. S. Sheen, H. Y. Chiou, C. M. Chu, and Y. F. Liaw. 2002. Long-term outcome after spontaneous HBeAg seroconversion in patients with chronic hepatitis B. Hepatology 35:1522-1527. [DOI] [PubMed] [Google Scholar]
- 9.Lindh, M., A. S. Andersson, and A. Gusdal. 1997. Genotypes, nt 1858 variants, and geographical origin of hepatitis B virus—large-scaled analysis using a new genotyping method. J. Infect. Dis. 175:1285-1293. [DOI] [PubMed] [Google Scholar]
- 10.Lindh, M., P. Horal, A. P. Dhillon, and G. Norkrans. 2000. Hepatitis B virus DNA levels, precore mutations, genotypes and histological activity in chronic hepatitis B. J. Viral Hepat. 7:258-267. [DOI] [PubMed] [Google Scholar]
- 11.Loeb, K. R., K. R. Jerome, J. Goddard, M. L. Huang, A. Cent, and L. Corey. 2000. High-throughput quantitative analysis of hepatitis B virus DNA in serum using the TaqMan fluorogenic detection system. Hepatology 32:626-629. [DOI] [PubMed] [Google Scholar]
- 12.Lok, A. S. F., C. L. Lai, P. C. Wu, E. K. Y. Leung, and T. S. Lam. 1987. Spontaneous hepatitis B e antigen to antibody seroconversion and reversion in Chinese patients with chronic hepatitis B virus infection. Gastroenterology 92:1839-1843. [DOI] [PubMed] [Google Scholar]
- 13.Lok, A. S. F., E. J. Heathcote, and J. H. Hoofnagle. 2001. Management of hepatitis B: 2000—summary of a workshop. Gastroenterology 120:1828-1853. [DOI] [PubMed] [Google Scholar]
- 14.Manesis, E. L., G. V. Papatheodoridis, and S. J. Hadziyannis. 2002. Serum HBV-DNA levels in inactive hepatitis B virus carriers. Gastroenterology 122:2092-2093. [DOI] [PubMed] [Google Scholar]
- 15.Martinot-Peignoux, M., N. Boyer, M. Colombat, R. Akremi, B. N. Pham, S. Ollivier, C. Castelnau, D. Valla, C. Degott, and P. Marcellin. 2002. Serum hepatitis B virus DNA levels and liver histology in inactive HBsAg carriers. J. Hepatol. 36:543-546. [DOI] [PubMed] [Google Scholar]
- 16.McMahon, B. J., P. Holck, L. Bulkow, and M. Smowball. 2001. Serologic and clinical outcomes of 1536 Alaska natives chronically infected with hepatitis B virus. Ann. Intern. Med. 135:759-768. [DOI] [PubMed] [Google Scholar]
- 17.Sung, J. J. Y., H. L. Y. Chan, M. L. Wong, C. H. Tse, S. C. H. Yuen, J. S. L. Tam, and N. W. Y. Leung. 2002. Relationship of clinical and virological factors with hepatitis activity in hepatitis B e antigen-negative chronic hepatitis B virus-infected patients. J. Viral Hepat. 9:229-234. [DOI] [PubMed] [Google Scholar]
- 18.ter Borg, F., F. J. W. ten Kate, H. T. M. Cuypers, A. Leentvaar-Kuijpers, J. Oosting, P. M. E. Wertheim-van Dillen, P. Honkoop, M. C. Rasch, R. A. de Man, J. van Hattum, R. A. F. M. Chamuleau, H. W. Reesink, and E. A. Jones. 1998. Relation between laboratory test results and histological hepatitis activity in individuals positive for hepatitis B surface antigen and antibodies to hepatitis B e antigen. Lancet 351:1914-1918. [DOI] [PubMed] [Google Scholar]