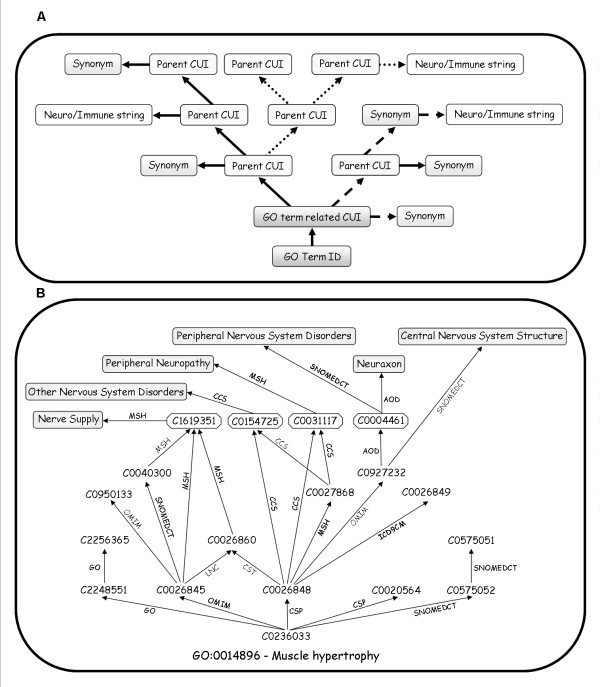
Figure 4.
The UMLStermFinder. (A) The underlying algorithms. A GO term is associated with a Unique Concept Identifier (CUI) using the UMLS classes file (MRSAT). The UMLS relations file (MRREL) is then used to identify CUI terms that are parental to the input GO terms, recursively traversing the DAG up to 3 levels up. All the aliases to any of these terms are also extracted. Each CUI found in the resulting sub-graph is associated with their corresponding strings, which are searched for any of the following: 'immune', 'neuro', 'inflam', 'brain', 'lymph' or 'nerve'. The outputs of the algorithm are the terms that contain these strings. Arrows of different types denote different sources from which relationships were asserted. (B) An example of how UMLStermFinder works. Using the GO term 'muscle hypertrophy' (GO:0014896) to search the UMLS ontologies, the corresponding concept identifier is found and used to find the concept's parents and synonyms (framed CUIs) of the initial concept and its parents. After all concepts are collected, they are searched for neural and/or immune-related string content (grey boxes). The UMLStermFinder finds concepts from several ontologies; by each arrow in the figure appears the name of the source ontology from which the relationship was asserted. A Perl script implementing the UMLStermFinder algorithm is freely available as Additional file 7.