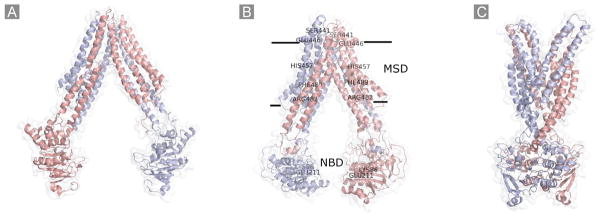
Fig. 4.
Schematic representation of the homology models of BCRP. A, the substrate-unbound nucleotide-free inward-facing open apo conformation based on the MsbA structure (PDB code 3B5W); B, the substrate-bound nucleotide-free inward-facing closed apo conformation based on the mouse P-gp structure (PDB code 3G60). The approximate locations of several amino acid residues in the MSD (Ser441, Glu446, His457, Phe489, and Arg482) or the NBD (Lys86 and Glu211) that could be important for substrate specificity and/or overall transport activity are indicated; C, the nucleotide-bound outward-facing conformation based on the Sav1866 structure (PDB code 2HYD). Two monomers in the BCRP dimer are shown in different colors.