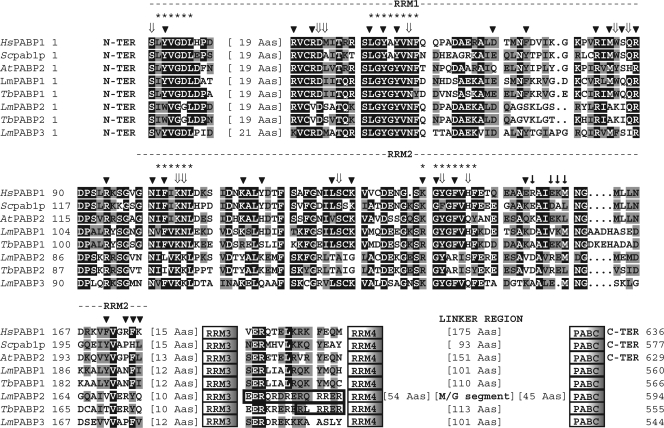
Fig. 1.
Sequence analysis of the L. major and T. brucei PABP homologues. Shown is a ClustalW alignment comparing the sequences from RRM1 and -2 of the human (Hs), yeast (Sc; S. cerevisiae), and plant (At; A. thaliana) PABP homologues with the various L. major/T. brucei proteins. The two RRMs are indicated, as well as the positions of RRM3 and -4, the linker region, and the PABC domain. Sequences between RRM3 and -4 where putative NLS were identified in the PABP2 orthologues are also shown. Within the aligned regions, amino acids identical in more than 60% of the sequences are highlighted in dark gray, while amino acids defined as similar, based on the BLOSUM 62 Matrix, in more than 60% of the sequences are shown in pale gray. When necessary, spaces were inserted within the various sequences (dots) to allow better alignment. The asterisks define the two RNP motifs. The arrowheads indicate amino acids that have been shown to interact with the RNA, while ⇓ highlights amino acids specifically involved in poly(A) recognition (18). ↓ symbols indicate the residues in yeast PABP RRM2 relevant to the interaction with eIF4G (based on reference 48). The location of the methionine/glycine-rich segment within the linker region from LmPABP2 is highlighted. The A. thaliana PABP2 homologue was included in the alignment because it is the nearest homologue to LmPABP1 within the various A. thaliana PABPs (not shown). For the GenBank/GeneDB accession numbers for the trypanosomatid PABP homologues, see Materials and Methods. The other GenBank accession numbers are as follows: HsPABP, P11940; Scpab1p, NP_011092; AtPABP2, P42731.