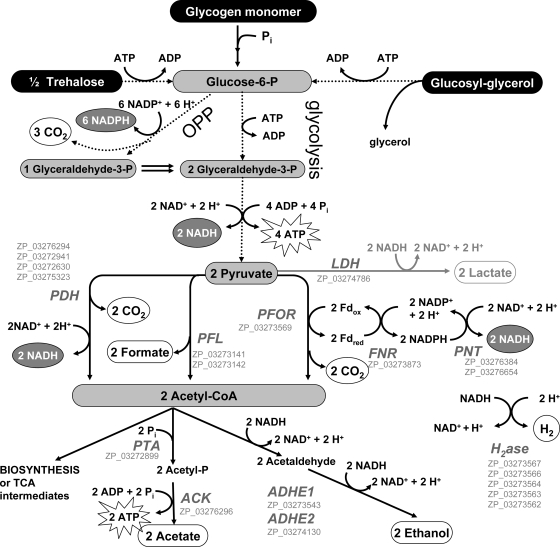
FIG. 5.
Proposed autofermentative scheme for A. maxima based on observed excreted products (white circles), previously observed substrates (black circles) (8), and identified enzymes present in the A. maxima genome (GenBank accession numbers [gray letters]). Pathways with multiple steps (glycolysis and oxidative pentose phosphate [OPP] pathways) are given as dotted lines. Lactate production is provided in gray because it was a minor end product for all experiments in this study. Enzyme abbreviations: LDH, lactate dehydrogenase; PDH, pyruvate dehydrogenase; PFL, pyruvate formate lyase; PFOR, pyruvate ferredoxin oxidoreductase; FNR, ferredoxin NADP reductase; PNT, pyridine NADH transhydrogenase; PTA, phosphotransacetylase; ACK, acetate kinase; ADHE, bifunctional aldehyde alcohol dehydrogenase; H2ase, hydrogenase. The enzymes required for the glycolysis and OPP pathways and the biosynthetic/degradation pathways for trehalose and glucosyl-glycerol were identified in the A. maxima genome (see the supplemental material for annotation of the glucosyl-glycerol, trehalose, and glycogen pathways).