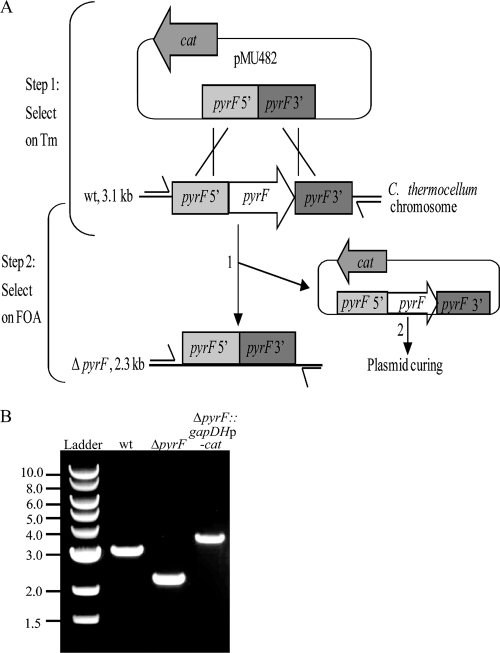
FIG. 1.
Creation of a C. thermocellum ΔpyrF strain. (A) Relevant features of the pyrF deletion vector, pMU482, are the positive marker for transformation (cat) and the flanking regions of homology that target deletion of pyrF (pyrF 5′ and pyrF 3′). The chromosomal pyrF locus contains the pyrF gene in the context of the flanking DNA. Step 1, plating on Tm allows selection of transformants harboring pMU482; step 2, plating on 5-FOA selects for deletion of pyrF from chromosome and insertion into pMU482 (numbered 1) and loss of pMU482 containing pyrF (numbered 2). Primers used for PCR are indicated by the one-sided arrows, and the sizes of the expected amplicons are shown to the left. (B) DNA gel showing colony PCR results using primers X02204 and X02205 on wild-type (lane 2), ΔpyrF (lane 3), and ΔpyrF::gapDHp-cat (lane 4) strains. The numbers on the left indicate the band sizes in kb for the NEB 1-kb ladder used as a marker (lane 1).