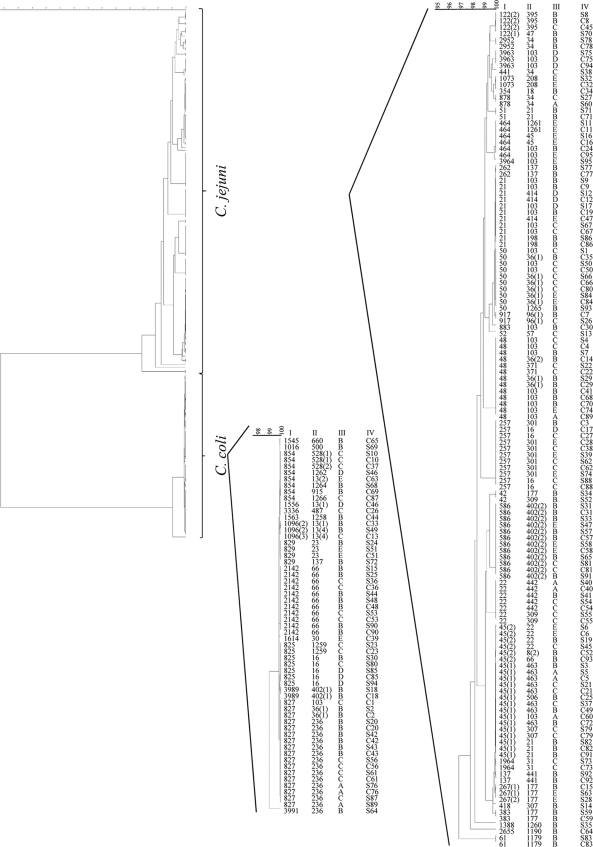
FIG. 1.
Clustering of the paired sample strains using the entire set of MLST target sequences. The unweighted-pair group method using average linkages (UPGMA) tree was applied in BioNumerics. The ST (I), flaB types (II), and suppliers' designations (III) are indicated. Paired samples (IV) are labeled with corresponding numbers for the cecum (C) and neck skin (S). The numbers in parentheses indicate identical STs that are variants based on differences that were observed within flanking regions of the sequence used for allele definition by PubMLST.