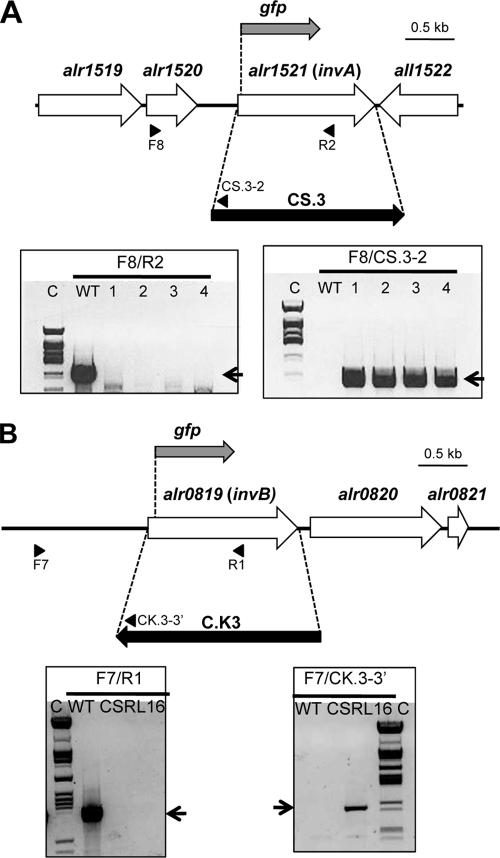
FIG. 1.
Schematic of gfp reporter fusions and insertional inactivation of the invA (A) and invB (B) genes. Strategies of insertion of gene cassettes to produce strains CSRL17 (invA) and CSRL16 (invB) and sites of insertion of the gfp gene to produce strains CSRL19 (invA-gfp) and CSRL18 (invB-gfp) are shown. The approximate positions of the primers used to check segregation of the mutant chromosomes by PCR are indicated in each case. PCR analysis of the genetic structure in the invA (A) and invB (B) regions in strains PCC 7120, CSRL17, and CSRL16 is also shown, with the indication of the primer pairs used for amplification. Electrophoresis gel lanes: C, λ DNA digested with ClaI; WT, reactions with DNA from strain PCC 7120; 1 to 4 in panel A, reactions with DNA from 4 different exconjugants; CSRL16 in panel B, reaction with DNA from strain CSRL16. The expected amplification products are as follows: for F8/R2, 1,630 bp (WT) and none in the mutant clones (note that the R2 primer is within the deleted DNA sequences); for F8/CS.3-2, none in the WT (the CS3-2 primer is within the CS.3 gene cassette) and 805 bp in the mutants; for F7/R1, 1,577 bp (WT) and none in CSRL16 (the R1 primer is within the deleted DNA sequences); for F7/CK.3-3′, none in the WT (the CK3-3′ primer is within the CK.3 gene cassette) and 995 bp in CSRL16.