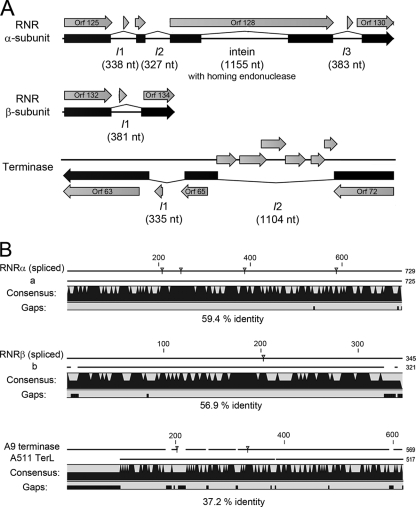
FIG. 7.
Identification of intervening sequences (IVS) in the A9 genome. (A) Splicing variants producing the most significant database hits were annotated. The putative intein was identified by Blastp. RNR, ribonucleotide reductase. (B) Alignment of the spliced proteins with homologous sequences which do not contain IVS (a, RNRα subunit of Granulicatella elegans ATCC 700633; b, RNRβ subunit of Enterococcus casseliflavus EC30; A511 TerL, large terminase of Listeria phage A511). Amino acid sequence identities and gaps in the alignment are indicated. Small triangles mark the putative processing sites.