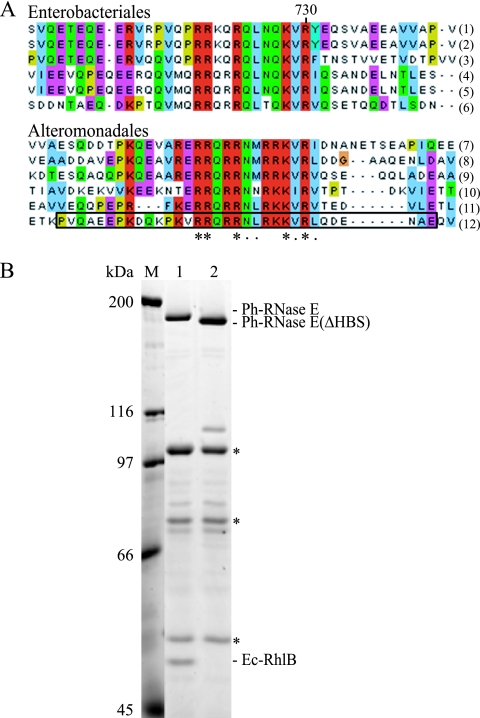
FIG. 6.
Identification of the RhlB binding site of Ph-RNase E. (A) Sequence alignment of the HBSs in homologs from the Enterobacteriales (top) and the corresponding regions in homologs from the Alteromonadales (bottom). The asterisks indicate residues that are conserved; the dots indicate residues that are similar. The homologs used in these alignments correspond to Escherichia coli (1), Shigella boydii Sb227 (2), Salmonella enterica subsp. enterica (3), Yersinia pseudotuberculosis IP 31758 (4), Erwinia carotovora subsp. atroseptica SCRI1043 (5), Yersinia pestis Antiqua (6), Shewanella sp. ANA-3 (7), Shewanella amazonensis SB2B (8), Shewanella sp. PV-4 (9), Colwellia psychrerythraea 34H (10), Pseudoalteromonas tunicata D2 (11), and Pseudoalteromonas haloplanktis TAC125 (12). The deletion of the region corresponding to the motif identified in Ph-RNase E in pSAB18 to create pSAB29 corresponds to the boxed sequence. (B) SDS-PAGE stained with Sypro orange. Immunopurifications were performed as shown in Fig. 2. Lane 1, pSAB18 (Ph-RNase E with FLAG); lane 2, pSAB29, which expresses a variant of Ph-RNase E deleted from residues 684 to 716 (boxed in panel A, last line). M, molecular mass markers. The asterisks correspond to the subunits of pyruvate dehydrogenase, which interacts adventitiously with the FLAG antibody.