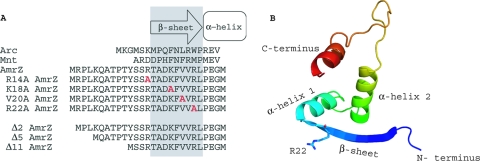
FIG. 1.
Alignment and predicted secondary structure of the putative ribbon-helix-helix transcriptional regulator AmrZ. (A) An amino acid alignment of the Arc-like DNA-binding domains of Arc (residues 1 to 18), Mnt (residues 1 to 15), and AmrZ (residues 1 to 27) reveals conserved residues in the DNA-binding β-sheet as well as the presence of the extended amino terminus. Residues in gray indicate the DNA-binding β-sheet. Residues to the left are part of the extended amino acid. Residues within the DNA-binding β-sheet that were targeted for site-specific mutagenesis are shown in red. (B) Predicted three-dimensional (3D) structure of AmrZ residues 13 to 80 provided by the secondary-structure prediction program pyMol. The major structural components (N and C termini, β-sheet, and α-helices) are indicated. The location of arginine-22 (R22) is also indicated, given the frequent references to the R22A mutant.